Abstract
Background
The seedling establishment is controlled by the programmed expression of sets of genes at the specific tissues of seed, abundance and environment. Plumule is an important part of the seed embryo and expresses the suits of genes to exert distinct functions during seed germination. Although rice genomic resources are available and developed rapidly, thousands of transcripts have not previously been located in the plumule of rice.
Objective
This study was performed to identify plumule-preferentially expressed (OsPluP) genes in rice and determine the expression profiles and functions of OsPluP genes.
Methods
We identified the OsPluP genes through Affymetrix microarray data. Meanwhile, qRT-PCR was performed to validate the expression pattern, also found that OsPluP genes were regulated by dark/light treatment. The cis-acting regulatory elements were analyzed in the promoters’ regions of OsPluP genes. The T-DNA mutant of the OsPluP seed was used to reveal the function in seed germination.
Results
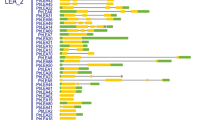
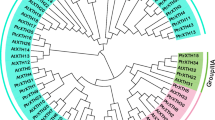
In this study, a genomic survey of OsPluP genes was performed, and we identified 88 OsPluP genes based on Affymetrix microarray data. The expression profiles of 88 OsPluP members in 24 representative tissues covering rice whole life cycle can be roughly classified into three major groups, suggesting functional divergence of OsPluP genes in seed germination. The microarray data, qRT-PCR, and promoter analysis results demonstrated that transcripts of more than half OsPluPs (54 genes) could be enhanced in the darkness and respond to phytohormone. Gene Ontology (GO)and Kyoto encyclopedia of genes and genomes (KEGG) analysis demonstrated that OsPluP and their co-expressed genes were highly enriched in fatty acid metabolism. Moreover, OsPluP82 T-DNA mutant seeds displayed short plumule length and storage lipid accumulation.
Conclusion
This study would enable the functions of OsPluP genes during seed germination and contribute to the goal of molecular regulatory networks that lay the foundation for further studies of seedling growth.








Similar content being viewed by others
Data availability
The expression profile of the OsPluP genes in various tissues of cultivar Minghui 63 was extracted from NCBI Gene Expression Omnibus GSE19024.
References
Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou JP, Grini PE, Colot V et al (2011) Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7:e1002014
Carroll SB (2001) Chance and necessity: the evolution of morphological complexity and diversity. Nature 409:1102–1109
Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49:311–343
Chepyshko H, Lai CP, Huang LM, Liu JH, Shaw JF (2012) Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: new insights from bioinformatics analysis. BMC Genom 13:309
Feitosa-Araujo E, de Souza CI, Florian A, da Fonseca-Pereira P, Condori Apfata JA, Heyneke E, Medeiros DB, Pires MV, Mettler-Altmann T, Neuhaus HE et al (2020) Downregulation of a mitochondrial NAD+ transporter (NDT2) alters seed production and germination in Arabidopsis. Plant Cell Physiol 61:897–908
Feussner I, Kuhn H, Wasternack C (2001) Lipoxygenase-dependent degradation of storage lipids. Trends Plant Sci 6:268–273
Finch-Savage WE, Bassel GW (2016) Seed vigour and crop establishment: extending performance beyond adaptation. J Exp Bot 67:567–591
Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19:485–494
Gong D, He F, Liu J, Zhang C, Wang Y, Tian S, Sun C, Zhang X (2022) Understanding of hormonal regulation in rice seed germination. Life (Basel) 12:1021. https://doi.org/10.3390/life12071021
Haas BJ, Delcher AL, Wortman JR, Salzberg SL (2004) DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20:3643–3646
Han C, Zhen S, Zhu G, Bian Y, Yan Y (2017) Comparative metabolome analysis of wheat embryo and endosperm reveals the dynamic changes of metabolites during seed germination. Plant Physiol Biochem 115:320–327
Heazlewood JL, Howell KA, Whelan J, Millar AH (2003) Towards an analysis of the rice mitochondrial proteome. Plant Physiol 132:230–242
Howell KA, Millar AH, Whelan J (2006) Ordered assembly of mitochondria during rice germination begins with pro-mitochondrial structures rich in components of the protein import apparatus. Plant Mol Biol 60:201–223
Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, Usadel B, Millar AH, Whelan J (2009) Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149:961–980
Huang J, Cai M, Long Q, Liu L, Lin Q, Jiang L, Chen S, Wan J (2014) OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgen Res 23:643–655
Kadota K, Ye J, Nakai Y, Terada T, Shimizu K (2006) ROKU: a novel method for identification of tissue-specific genes. BMC Bioinform 7:294
Karssen CM, Brinkhorst-van der Swan DL, Breekland AE, Koornneef M (1983) Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157:158–165
Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13:571–577
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li QF, Wang JD, Xiong M, Wei K, Zhou P, Huang LC, Zhang CQ, Fan XL, Liu QQ (2018) iTRAQ-based analysis of proteins co-regulated by brassinosteroids and gibberellins in rice embryos during seed germination. Int J Mol Sci 19:3460
Li Z, Liang Y, Yuan Y, Wang L, Meng X, Xiong G, Zhou J, Cai Y, Han N, Hua L et al (2019) OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localization. Mol Plant 12:1143–1156
Liew LC, Narsai R, Wang Y, Berkowitz O, Whelan J, Lewsey MG (2020) Temporal tissue-specific regulation of transcriptomes during barley (Hordeum vulgare) seed germination. Plant J 101:700–715
Liu X, Bai X, Wang X, Chu C (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164:969–979
Lu Z, Huang X, Ouyang Y, Yao J (2013) Genome-wide identification, phylogenetic and co-expression analysis of OsSET gene family in rice. PLoS ONE 8:e65426
Martinez-Hernandez A, Lopez-Ochoa L, Arguello-Astorga G, Herrera-Estrella L (2002) Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol 128:1223–1233
Miro B, Longkumer T, Entila FD, Kohli A, Ismail AM (2017) Rice seed germination underwater: morpho-physiological responses and the bases of differential expression of alcoholic fermentation enzymes. Front Plant Sci 8:1857
Miura K, Lin SY, Yano M, Nagamine T (2002) Mapping quantitative trait loci controlling low temperature germinability in rice (Oryza sativa L.). Breed Sci 51:293–299
Narsai R, Law SR, Carrie C, Xu L, Whelan J (2011) In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol 157:1342–1362
Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, Lister R, Lewsey MG, Whelan J (2017) Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol 18:172
Nayak RR, Kearns M, Spielman RS, Cheung VG (2009) Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res 19:1953
Ngai N, Tsai FY, Coruzzi G (1997) Light-induced transcriptional repression of the pea AS1 gene: identification of cis-elements and transfactors. Plant J 12:1021–1034
Niu Y, Wu GZ, Ye R, Lin WH, Shi QM, Xue LJ, Xu XD, Li Y, Du YG, Xue HW (2009) Global analysis of gene expression profiles in Brassica napus developing seeds reveals a conserved lipid metabolism regulation with Arabidopsis thaliana. Mol Plant 2:1107–1122
Ouyang Y, Huang X, Lu Z, Yao J (2012) Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genom 13:100
Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y (2013) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41:D1206-1213
Schmidt MA, Herman EM (2018) Characterization and functional biology of the soybean aleurone layer. BMC Plant Biol 18:354
Shi J, Zhou H, Liu X, Wang N, Xu Q, Yan G (2021) Correlation analysis of the transcriptome and metabolome reveals the role of the flavonoid biosynthesis pathway in regulating axillary buds in upland cotton (Gossypium hirsutum L.). Planta 254:7. https://doi.org/10.1007/s00425-021-03597
Topham AT, Taylor RE, Yan D, Nambara E, Johnston IG, Bassel GW (2017) Temperature variability is integrated by a spatially embedded decision-making center to break dormancy in Arabidopsis seeds. Proc Natl Acad Sci USA 114:6629–6634
Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D et al (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135:1206–1220
Vishal B, Kumar PP (2018) Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front Plant Sci 9:838
Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J (2010) A dynamic gene expression atlas covering the entire life cycle of rice. Plant J 61:752–766
Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J et al (2010b) A dynamic gene expression atlas covering the entire life cycle of rice. Plant J 61:752–766
Wang L, Qin L, Liu W, Zhang D, Wang Y (2014) A novel ethylene-responsive factor from Tamarix hispida, ThERF1, is a GCC-box- and DRE-motif binding protein that negatively modulates abiotic stress tolerance in Arabidopsis. Physiol Plant 152:84–97
Wei T, He Z, Tan X, Liu X, Yuan X, Luo Y, Hu S (2015) An integrated RNA-Seq and network study reveals a complex regulation process of rice embryo during seed germination. Biochem Biophys Res Commun 464:176–181
Yang XC, Hwa CM (2008) Genetic and physiological characterization of the OsCem mutant in rice: formation of connected embryos with multiple plumules or multiple radicles. Heredity (edinb) 101:239–246
Yang Y, Li Y, Wu C (2013) Genomic resources for functional analyses of the rice genome. Curr Opin Plant Biol 16:157–163
Yang W, Lu Z, Xiong Y, Yao J (2017) Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop J 5:21–31
Yi X, Du Z, Su Z (2013) PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res 41:W98-103
Zhang N, Yu H, Yu H, Cai Y, Huang L, Xu C, Xiong G, Meng X, Wang J, Chen H et al (2018) A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell 30:1461–1475
Zhao X, Huang J, Yu H, Wang L, Xie W (2010) Genomic survey, characterization and expression profile analysis of the peptide transporter family in rice (Oryza sativa L). BMC Plant Biol 10:92. https://doi.org/10.1186/1471-2229-10-92
Zhou Z, Wang MJ, Zhao ST, Hu JJ, Lu MZ (2010) Changes in freezing tolerance in hybrid poplar caused by up- and down-regulation of PtFAD2 gene expression. Transgen Res 19:647–654
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant no. 32160699, 32060587 and 31660046), Guizhou Province Natural Science Foundation [QKHJC-ZK[2022]ZD032]. The Joint Fund of the National Natural Science Foundation of China and the Karst Science Research Center of Guizhou province (Grant no. U1812401). Guizhou Educational project Qianjiaohe ([2021]309).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declared no conflict of interest with respect to the research, authorship, or publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, G., Wu, Z., Zhai, L. et al. Identification and co-expression network analysis of plumule-preferentially expressed genes in Oryza sativa. Genes Genom 45, 319–336 (2023). https://doi.org/10.1007/s13258-023-01366-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-023-01366-w