Key message
This study identified candidate genes related to fruit yield for an emerging medicinal crop, weeping forsythia.
Abstract
Background
The genetic basis of crop yield is an agricultural research hotspot. Identifying the genes related to yield traits is the key to increase the yield. Weeping forsythia is an emerging medicinal crop that currently lacks excellent varieties. The genes related to fruit yield in weeping forsythia have not been identified.
Objective
Thus, we aimed to screen the candidate genes related to fruit yield of weeping forsythia by using genome-wide association analysis.
Methods
Here, 60 samples from the same field and source of weeping forsythia were collected to identify its yield-related candidate genes. Association analysis was performed on the variant loci and the traits related to yield, i.e., fruit length, width, thickness, and weight.
Results
Results from admixture, neighbor-joining, and kinship matrix analyses supported the non-significant genetic differentiation of these samples. Significant association was found between 2 variant loci and fruit length, 8 loci and fruit width, 24 loci and fruit thickness, and 13 loci and fruit weight. Further search on the 20 kb up/downstream of these variant loci revealed 1 gene related to fruit length, 16 genes related to fruit width, 12 genes related to fruit thickness, and 13 genes related to fruit weight. Among which, 4 genes, namely, WRKY transcription factor 35, salicylic acid-binding protein, auxin response factor 6, and alpha-mannosidase were highly related to the fruit development of weeping forsythia.
Conclusion
This study identify four candidate genes related to fruit development, which will provide useful information for the subsequent molecular-assisted and genetic breeding of weeping forsythia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genetic basis of crop yield is an agricultural research hotspot (Luo et al. 2021; Achary et al. 2020). Genes affecting crop yield must be identified to reveal the genetic basis and facilitate the increase in crop yield through genetic engineering and hybridization or selective breeding (Gu et al. 2020). Genome-wide association study (GWAS) and quantitative trait locus (QTL) analysis are the most efficient methods for detecting yield-related genes in staple crops, such as wheat, maize, rice, and cotton (Alemu et al. 2020; Su et al. 2020; Wang et al. 2020; Zhang et al. 2020). However, the yield-related genes of emerging crops are difficult to identify by using GWAS or QTL analysis due to the lack of genomic information. With its further development, the cost of the development of third-generation sequencing has dropped exponentially. Many genome sequences of emerging crops have been published and thus serve as a basis to reveal the genetic mechanism underlying their yield.
Weeping forsythia (Forsythia suspensa (Thunb.) Vahl) is a perennial plant of the genus Forsythia in the Oleaceae family. The main medicinal ingredients of this fruit are forsythin and forsythiaside A, which have broad-spectrum antibacterial and antiviral effects (Yan et al. 2016). Its fruit extracts are mostly incorporated in Chinese traditional medicines for influenza treatment (Wang et al. 2018). Lianhua Qingwen capsules containing this fruit extract can remarkably alleviate the symptoms of COVID-19 (Hu et al. 2020). The weeping forsythia fruits currently sold in the market have been harvested from wild populations; however, their yield and quality have declined due to recent “robbing green” phenomenon (Wang et al. 2018). The market demand for weeping forsythia has increased greatly due to its huge medicinal value, but the current supply from wild resources is insufficient (Li et al. 2020a). Large-scale artificial planting has been initiated in Henan, Shanxi, Shaanxi, Shandong, Hebei, and other provinces in China. However, most of the current cultivated populations are genetically mixed, and no high-yield varieties are applied for production. The shape and weight of field weeping forsythia fruits vary greatly and therefore affect its yield. The current yield is low at only 2250–3750 kg per hm2. Hence, additional planting areas are needed to meet the market demand.
Crop yield can be increased by enhancing variety, cultivation techniques, and environmental conditions (Zhu et al. 2015). Among which, variety improvement is the most important; identifying the genes related to crop yield greatly accelerates breeding speed. Fruit size is a quantitative trait controlled by multiple genes. In Cucumis sativus L. (Yang et al. 2013; Wei et al. 2016) and Citrullus lanatus L. (Ren et al. 2014; Dou et al. 2018), the fruit size is controlled by multiple genes. In Solanum lycopersicum L., 41 gene loci are related to fruit traits, and 17 of which are related to fruit size (Phan et al. 2019).
Linkage and association analyses are the main screening methods for quantitative trait genes (Sahu et al. 2015). Linkage analysis creates a mapping population through parental crossing and then maps the genes that control quantitative traits to the genome. This method has achieved great success in locating traits that conform to Mendelian inheritance (Ma et al. 2020). Association analysis does not require a mapping group and can use natural or cultivated groups as research materials. Natural groups undergo multiple rounds of genetic recombination, and linkage disequilibrium decay (LDD) exists within a short distance, thus allowing the precise positioning of genes (Li et al. 2020b). Association analysis is an important method to identify genes that control quantitative traits in crops (Daba et al. 2020). This technique is classified into genome-wide (GWAS) and candidate gene-based association analysis; the former is suitable for locating genes with unknown function. In Zea mays ssp. mays L., GWAS identified 25 known genes related to flowering time and photoperiod sensitivity and 37 new genes related to days to tassel, anthesis-silk interval, and photoperiod sensitivity (Li et al. 2020b). In Triticum aestivum L., 4 genes associated with plant dwarfing were determined using this method (Daba et al. 2020). In Glycine max (Linn.) Merr., 23 genome segments related to agronomic traits, such as yield, maturity days, seed oil content, seed protein, and 100-seed weight were recognized by GWAS (Bruce et al. 2020). These studies confirmed that GWAS effectively identifies genes that control quantitative traits in crops.
GWAS usually requires a large sample size for detection to improve the statistical power and recognize many candidate genes (Wang and Xu 2019). However, the use of at least several hundred samples requires enormous costs. Here, the yield-related candidate genes of weeping forsythia, an emerging medicinal crop, were identified by using a small number of cultivated samples from the same field and source. Although these fruits are from the same field and source, they still exhibit differences in shape and weight.
Weeping forsythia is a woody plant that takes 3 years from seeding to fruiting. Therefore, a long breeding cycle is required through conventional breeding. Effective methods must be urgently adopted to improve the breeding speed of the high-yielding varieties of this emerging medicinal crop. The identified genes related to the fruit size of weeping forsythia will serve as the foundation for molecular-assisted and genetic engineering breeding to greatly improve its breeding efficiency. The recently published genome sequence of weeping forsythia (Li et al. 2020a) was used as the basis.
In this study, weeping forsythia samples cultivated in the field were selected. Association analysis was performed based on the phenotypic data of fruit shape and weight and variant loci from genotyping by sequencing (GBS) to screen the candidate genes related to fruit yield. The findings will be useful for the subsequent molecular-assisted and genetic engineering breeding of weeping forsythia.
Materials and methods
Sampling and phenotypic measurement
Immature fruits of weeping forsythia, known as ‘Qingqiao,’ were harvested in July to extract their medicinal ingredients. Fresh and healthy fruits (Figs. 1 and S1) were collected from 60 seedlings of 5-year-old plants. All the individual plants are located in the same field in Hexi village (34.03° N, 111.03° E), Sanmenxia, China. Ten fruits were collected from each individual. The average length, width, thickness and weight of 10 fruits per plant were measured using a vernier caliper. Pearson correlation analysis was applied for the phenotypic traits using cor.test in R, where r > 0.7 is taken as the threshold of correlation. Fresh leaves were collected from the corresponding 60 seedlings and immediately stored in silica gel for subsequent DNA extraction.
Library construction and genotyping
DNA extractions for each individual were performed on 30 mg of silica gel dried leaf material using CTAB methodology (Doyle and Doyle 1987). Genomic DNA was detected using the following methods to ensure the quality of library construction. (1) Agarose gel electrophoresis was performed to select the samples with the main band and prevent degradation and RNA contamination. (2) Nanodrop 2000 (Thermo Fisher Scientific, MA, USA) was used to select samples with an OD260/280 ratio between 1.8 and 2.2. (3) Qubit 3.0 (Thermo Fisher Scientific, MA, USA) was employed to detect samples with DNA concentration greater than 50 ng/µl and total amount greater than 2 µg. The qualifying DNA was randomly interrupted and then prepared following the steps of terminal repair, (A) tail addition, sequencing adaptor addition, purification, and polymerase chain reaction (PCR) amplification. After the library was constructed, Qubit 3.0 (Thermo Fisher Scientific, MA, USA) was applied for preliminary quantification and library dilution. Agilent 2100 (Agilent, CA, USA) was employed to detect the size of inserted fragments of the library. After the inserted fragments reached the expected size, qPCR was used to accurately quantify the effective concentration of the library to ensure its quality. The qualified libraries were sequenced on an Illumina NovaSeq system (Illumina, Inc., CA, USA) following the manufacturer's protocols.
Fastp 0.20.1 (Chen et al. 2018) was used for data quality control and filtering with the following standards: reads with polyG and polyX at the tail, N number greater than 5, base mass lower than 15, and ratio higher than 40%. After filtering, reads less than 15 in length were removed. The filtered reads were mapped onto the reference genome of weeping forsythia by using BWA 0.7.17 (Li and Durbin 2009). Variant loci were identified by using the GATK 4.1.8.1 (McKenna et al. 2010) and then annotated using snpEff 4.3 (Cingolani et al. 2012).
Population structure analysis
GATK was used to filter variant loci with the following filter parameters to obtain high quality SNP and Indel: QD < 2.0, FS > 60.0, MQ < 20.0, MQRankSum < − 12.5, and ReadPosRankSum < − 8.0. Vcftools 0.1.16 (Danecek et al. 2011) was applied to eliminate loci with MAF less than 3% and missing more than 0.2. The remaining loci were used for population structure analysis and GWAS.
Principal components analysis (PCA), admixture analysis, kinship relation analysis, and phylogenetic inferences were employed to infer the genetic relationships of all weeping forsythia samples. PCA was run using GCTA 1.93.2 (Yang et al. 2011). ADMIXTURE version 1.3 (Alexander et al. 2009) was used to investigate the maximum likelihood of the ancestry of each individual with co-ancestry clusters ranging from 1 to 8. Optimal clustering number was decided according to the cross validation (CV) error value. Phylogenetic relationships of the weeping forsythia samples were constructed using neighbor-joining method in PHYLIP 3.696 program (Felsenstein 2005). Kinship relation was analyzed using PLINK 1.90b6.12 (Purcell et al. 2007).
GWAS analysis
Given that the previous weeping forsythia genome (Li et al. 2020a) was not assembled at the chromosome level, many contigs were artificially combined into several chromosomes for analysis and data presentation. GWAS association analyses were performed on the merged files using EMMAX (Zhou and Stephens 2012) and FaST-LMM 0.4.6 (Lippert et al. 2011) with default parameters. The value of 1/variant loci (340,933) number was used as a threshold for screening variant loci associated with fruit traits. Genotyping was conducted on the variant loci and phenotypic data. Owing to the self-incompatibility of weeping forsythia (Qiao et al. 2020), the candidate genes related to fruit traits were searched on the 20 kb up/downstream of variant loci.
Results
Phenotypic traits of fruits
Four phenotypic traits, namely, fruit length, width, thickness, and weight, were measured (Table S1) for 10 weeping forsythia fruits. The weeping forsythia seedlings exhibited the following features: fruit length ranging 14.62–30.39 mm, fruit width ranging 8.30–14.86 mm, fruit thickness ranging 6.70–12.96 mm, and fruit weight ranging 0.38–1.71 g (Table S1, Fig. 2). The fruit length of weeping forsythia was correlated with the width (r = 0.510) and thickness (r = 0.471), but the correlation coefficients were not significant. Fruit size indexes such as length (r = 0.725), width (r = 0.847), and thickness (r = 0.821) were significantly correlated with fruit weight. According to the correlation coefficient, the fruit width and thickness significantly contributed to fruit weight.
Identification and distribution pattern of variant loci
The GBS of all samples yielded 313,066,384 raw reads with an average of 0.87 × sequencing depth. A total of 313,037,766 clean reads remained after filtering. An average of 5,217,296 reads per sample were mapped on the genome of weeping forsythia, and the average coverage of each sample in the genome reached 7.05%. A total of 3,586,221 SNPs, 399,021 inserts, and 226,432 deletions were recorded after variant locus detection. According to the definition of snpEFF, 96.79% of these loci belong to modifier variation, 1.66% belong to moderate variation, 1.12% belong to low variation, and only 0.43% belong to high variation. After further filtration by GATK and vcftools, the remaining 340,933 variant loci were used for subsequent GWAS and population structure analyses.
Population genetic structure
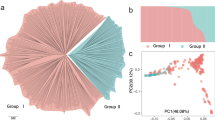
Admixture (Alexander et al. 2009) was used to examine the population genetic structure of 60 weeping forsythia samples based on all variant loci. An optimal clustering at K = 1 groups was confirmed based on the CV error rate (Fig. S2). Admixture does not support the genetic structural stratification of these weeping forsythia samples. The NJ tree (Fig. 3a) and Kinship matrix (Fig. 3b) did not verify significant genetic differentiation in the 60 samples. However, a genetic close relationship was observed for F34/F36, F35/F38, and F29/F30. This result was also supported by the PCA analysis (Fig. 3c).
Population genetic structure of weeping forsythia. A, Phylogenetic tree of all individuals of weeping forsythia were constructed using the neighbor-joining method in the program PHYLIP. B, Each point represents an individual genotype. The amount of variation explained by PC1 and PC2 is 3.62% and 3.42%. C, Kinship matrix of all individuals of weeping forsythia was constructed using PLINK
Characterization of candidate genes of fruit shape and weight traits
GWAS analysis with 340,933 variant loci was performed for the fruit traits, namely, length, width, thickness, and weight. Several variant loci passed the threshold line (− log10(p) > 5.533) and showed significant association with fruit length (2), fruit width (8), fruit thickness (24), and fruit weight (13) (Figs. 4 and S3-6). Given that weeping forsythia is a self-incompatible plant, a genetic distance of 20 kb was used to screen the candidate genes strongly related to its fruit traits. The results showed that 2 candidate gene was related to fruit length, 17 were related to fruit width, 44 were related to fruit thickness, and 17 were related to fruit weight (Table S2). On the basis of gene annotation and previous studies on gene function, most of these genes had unknown function or were unrelated to fruit development. Only 4 genes might be related to the fruit development of weeping forsythia (Table 1). In particular, WRKY transcription factor 35 (WRKY35, EVM0030555) and salicylic acid-binding protein 2 (SBP2, EVM0012993) were related to fruit width and thickness, and auxin response factor 6 (ARF6, EVM0018222), and alpha-mannosidase (EVM0029771) were related to fruit thickness (Table 1).
Discussion
Increasing crop yield is an agricultural production research hotspot (Dreccer et al. 2019). Variety improvement is an important way to increase the yield of cereal and industrial crops (Parry and Hawkesford 2012; Wan 2018). Identifying the genes related to yield traits is the key to this strategy. The genes can be used as molecular markers for offspring selection in the selective and cross breeding or target genes for genetic engineering breeding (Gu et al. 2020). This technology is especially important for woody crops with long breeding cycles (Rehman et al. 2020). GWAS can identify the target genes for woody crop research (De la Torre et al. 2019) and usually requires a large sample size. With small samples, identifying the target gene is difficult because of the lack of statistical power and the possibility of false positive results (Wang and Xu 2019).
Weeping forsythia is an emerging medicinal crop that currently lacks excellent varieties for production and cultivation. Seedlings in production are generated from the propagated seeds of natural populations. The fruits have different traits, leading to low production yield. Although reproduction can be achieved through cutting or tissue culture of superior individuals, these methods are limited. Identifying its yield-related genes is the key for the variety improvement of weeping forsythia.
In this study, all weeping forsythia seedlings were obtained the same field and source, Yuncheng, Shanxi, China. The fruits were genetically similar as confirmed by the population genetic structure analysis (Fig. 3). No genetic stratification was found, and only a few individuals showed genetic similarities, such as F34/F36, F35/F38, and F29/F30. Given that all seedlings were propagated from seeds and the species has self-incompatibility characteristics (Qiao et al. 2020), the genetically similar fruits still exhibited significantly different traits (Fig. 2). This finding provides an opportunity to use small samples to identify candidate genes related to fruit yield. Given the small sample size, two methods were used for analysis, namely, EMMAX and FASTLMM. A relatively high threshold was also applied to increase the statistical power.
Multiple variant loci related to fruit length, width, thickness, and weight were identified by EMMAX and FASTLMM. Further search on the upstream and downstream genes of these variant loci revealed multiple candidate genes related to fruit traits. Many gene functions of weeping forsythia are unknown because only a few studies were conducted on the gene functions of Oleaceae (Table S2). According to the functional annotations of genes from other parallel species, most of the identified candidate genes may not be related to fruit development (Table S2). However, the relationship between these genes and the fruit development of weeping forsythia needs further verification. Only the following four genes are most likely to be related to fruit development.
SBP2 (EVM0012993) is a 29 kDa protein that catalyzes the conversion of methyl salicylic acid into salicylic acid (SA) (Tripathi et al. 2010). Transgenic studies proved that the transfer of SBP2 gene can remarkably increase the endogenous SA content of plants (Guan et al., 2019). SA can also improve plant adaptability to biotic and abiotic stresses by activating plant defense systems (Tiwari et al. 2017). A recent study on pomegranate found that SA contributes to fruit development and could improve fruit yield and quality (García-Pastor et al. 2020). The present results suggested that SBP2 gene is related to the fruit width and thickness of weeping forsythia and thus contribute to its fruit size.
ARF6 is a member of the ARF gene family and encodes a transcription factor that combines with auxin-responsive promoter elements to regulate the expression of auxin response genes. Auxin is an important plant hormone involved in flower fertilization, fruit setting, and fruit formation and development (De Jong et al. 2009). This hormone regulates cell division and expansion during fruit development to control the fruit size (Devoghalaere et al. 2012). In the present study, ARF6 was significantly related to the fruit thickness of weeping forsythia. Recent reports on ARF gene family members ARF5 and ARF9 in tomato suggested that this gene family can affect fruit size by regulating cell division and expansion (De Jong et al. 2015; Liu et al. 2018).
WRKY35 (EVM0030555) is a transcription factor belonging to WRKY family that regulates plant growth and development and plays important roles in fruit development, such as fruit ripening (Cheng et al. 2016), fruit color (Wang et al. 2017), and fruit disease resistance (Deng et al. 2020). Here, WRKY35 was found to be possibly related to the fruit size of weeping forsythia. Alpha-mannosidase (EVM0029771) was also associated with fruit size, but previous reports have confirmed its relation to fruit ripening and softening (Irfan et al. 2016). Whether these identified genes are related to fruit development or have unknown functions related to fruit yield must be verified by transgenic experiments.
In this study, the seedlings of weeping forsythia from the same field and source were used as subjects. Owing to this plant’s self-incompatibility, its fruits are genetically similar but have distinctly different traits. This characteristic allows the use of small sample size to identify candidate genes related to its fruit yield. However, the identified genes still need further functional verification. The findings serve as an important basis for the breeding of high-yield varieties of this emerging medicinal crop.
References
Achary VMM, Sheri V, Manna M, Panditi V, Borphukan B, Ram B, Agarwal A, Fartyal D, Teotia D, Masakapalli SK, Agrawal PK, Reddy MK (2020) Overexpression of improved EPSPS gene results in field level glyphosate tolerance and higher grain yield in rice. Plant Biotechnol J 18:2504–2519
Alemu A, Feyissa T, Tuberosa R, Maccaferri M, Sciara G, Letta T, Abeyo B (2020) Genome-wide association mapping for grain shape and color traits in Ethiopian durum wheat (Triticum turgidum ssp. durum). Crop J 8:757–768
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Bruce RW, Torkamaneh D, Grainger CM, Belzile F, Eskandari M, Rajcan I (2020) Haplotype diversity underlying quantitative traits in Canadian soybean breeding germplasm. Theor Appl Genet 133:1967–1976
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890
Cheng Y, Ahammed GJ, Yu J, Yao Z, Ruan M, Ye Q, Li Z, Wang R, Feng K, Zhou G, Yang Y, Diao W, Wan H (2016) Putative WRKYs associated with regulation of fruit ripening revealed by detailed expression analysis of the WRKY gene family in pepper. Sci Rep 6:39000
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92
Daba SD, Tyagi P, Brown-Guedira G, Mohammadi M (2020) Genome-wide association study in historical and contemporary U.S. winter wheats identifies height-reducing loci. Crop J 8:243–251
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
De Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57:160–170
De Jong M, Wolters-Arts M, Schimmel BCJ, Stultiens CLM, de Groot PFM, Powers SJ, Tikunov YM, Bovy AG, Mariani C, Vriezen WH, Rieu I (2015) Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J Exp Bot 66:3405–3416
De la Torre AR, Wilhite B, Neale DB (2019) Environmental genome-wide association reveals climate adaptation is shaped by subtle to moderate allele frequency shifts in loblolly pine. Genome Biol Evol 11:2976–2989
Deng B, Wang W, Ruan C, Deng L, Yao S, Zeng K (2020) Involvement of CsWRKY70 in salicylic acid-induced citrus fruit resistance against Penicillium digitatum. Hortic Res 7:20157
Devoghalaere F, Doucen T, Guitton B, Keeling J, Payne W, Ling TJ, Ross JJ, Hallett IC, Gunaseelan K, Dayatilake GA, Diak R, Breen KC, Tustin DS, Costes E, Chagné D, Schaffer RJ, David KM (2012) A genomics approach to understanding the role of auxin in apple (Malus x domestica) fruit size control. BMC Plant Biol 12:7
Dou J, Zhao S, Lu X, He N, Zhang L, Ali A, Kuang H, Liu W (2018) Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theor Appl Genet 131:947–958
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dreccer MF, Molero G, Rivera-Amado C, John-Bejai C, Wilson Z (2019) Yielding to the image: how phenotyping reproductive growth can assist crop improvement and production. Plant Sci 282:73–82
Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 36. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
García-Pastor ME, Zapata PJ, Castillo S, Martínez-Romero D, Guillén F, Valero D, María S (2020) The effects of salicylic acid and its derivatives on increasing pomegranate fruit quality and bioactive compounds at harvest and during storage. Front Plant Sci 11:668
Gu QS, Ke HF, Liu ZW, Lv X, Sun ZW, Zhang M, Chen LT, Yang J, Zhang Y, Wu LQ, Li ZK, Wu JH, Wang GN, Meng CS, Zhang GY, Wang XF, Ma ZY (2020) A high-density genetic map and multiple environmental tests reveal novel quantitative trait loci and candidate genes for fibre quality and yield in cotton. Theor Appl Genet 133:3395–3408
Guan C, Wang C, Li Q, Ji J, Wang G, Jin C, Tong Y (2019) LcSABP2, a salicylic acid binding protein 2 gene from Lycium chinense, confers resistance to triclosan stress in Nicotiana tabacum. Ecotoxicol Environ Saf 183:109516
Hu K, Guan W, Bi Y, Zhang W, Li L, Zhang B, Liu Q, Song Y, Li X, Duan Z, Zheng Q, Yang Z, Liang J, Han M, Ruan L, Wu C, Zhang Y, Jia Z, Zhong N (2020) Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine 85:153242
Irfan M, Ghosh S, Meli VS, Kumar A, Kumar V, Chakraborty N, Chakraborty S, Datta A (2016) Fruit ripening regulation of α-mannosidase expression by the MADS box transcription factor RIPENING INHIBITOR and ethylene. Front Plant Sci 7:10
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li LF, Cushman SA, He YX, Li Y (2020a) Genome sequencing and population genomics modeling provide insights into local adaptation of weeping forsythia. Hortic Res 7:130
Li ZW, Liu XG, Xu XJ, Liu JC, Sang ZQ, Yu KC, Yang YX, Dai WS, Jin X, Xu YB (2020b) Favorable haplotypes and associated genes for flowering time and photoperiod sensitivity identified by comparative selective signature analysis and GWAS in temperate and tropical maize. Crop J 8:227–242
Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D (2011) FaST linear mixed models for genome-wide association studies. Nat Methods 8:833–835
Liu S, Zhang Y, Feng Q, Qin L, Pan C, Lamin-Samu AT, Lu G (2018) Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Sci Rep 8:2971
Luo QL, Zheng Q, Hu P, Liu LQ, Yang GT, Li HW, Li B, Li ZS (2021) Mapping QTL for agronomic traits under two levels of salt stress in a new constructed RIL wheat population. Theor Appl Genet 34:171–189
Ma FY, Du J, Wang OC, Wang H, Zhao BB, He GH, Yang ZL, Zhang T, Wu RH, Zhao FM (2020) Identification of long-grain chromosome segment substitution line Z744 and QTL analysis for agronomic traits in rice. J Integr Agric 19:1163–1169
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Parry MAJ, Hawkesford MJ (2012) An integrated approach to crop genetic improvement. J Integr Plant Biol 54:250–259
Phan NT, Trinh LT, Rho MY, Park TS, Kim OR, Zhao J, Kim HM, Sim SC (2019) Identification of loci associated with fruit traits using genome-wide single nucleotide polymorphisms in a core collection of tomato (Solanum lycopersicum L.). Sci Hortic 243:567–574
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Qiao Y, Cao Y, Jia M, Wang Y, He J, Zhang X, Wang W, Song Y (2020) Research on flower buds growth development and pollination habits of Forsythia suspensa heterostyly. Acta Hortic Sin 47:699–707
Rehman F, Gong HG, Li Z, Zeng SH, Yang TS, Ai PY, Pan LZ, Huang HW, Wang Y (2020) Identification of fruit size associated quantitative trait loci featuring SLAF based high-density linkage map of goji berry (Lycium spp.). BMC Plant Biol 20:474
Ren Y, McGregor C, Zhang Y, Gong G, Zhang H, Guo S, Sun H, Cai W, Jie Zhang J, Xu Y (2014) An integrated genetic map based on fourmapping populations and quantitative trait loci associated with economically important traits in watermelon (Citrullus lanatus). BMC Plant Biol 14:33
Sahu H, Amadabade J, Dhiri N (2015) Association mapping: a useful tool. Int J Plant Sci 10:85–94
Su JJ, Wang CX, Ma Q, Zhang A, Shi CH, Liu JJ, Zhang XL, Yang DL, Ma XF (2020) An RTM-GWAS procedure reveals the QTL alleles and candidate genes for three yield-related traits in upland cotton. BMC Plant Biol 20:416
Tiwari S, Lata C, Chauhan PS, Prasad V, Prasad M (2017) A functional genomic perspective on drought signalling and its crosstalk with phytohormone-mediated signalling pathways in plants. Curr Genomics 18:469–482
Tripathi D, Jiang YL, Kumar D (2010) SABP2, a methyl salicylate esterase is required for the systemic acquired resistance induced by acibenzolar-S-methyl in plants. FEBS Lett 584:3458–3463
Wan J (2018) Genetic crop improvement: a guarantee for sustainable agricultural production. Engineering 4:431–432
Wang MY, Xu SZ (2019) Statistical power in genome-wide association studies and quantitative trait locus mapping. Heredity 123:287–306
Wang L, Zhang XL, Wang L, Tian Y, Jia N, Chen S, Shi NB, Huang X, Zhou C, Yu Y, Zhang ZQ, Pang XQ (2017) Regulation of ethylene-responsive SlWRKYs involved in color change during tomato fruit ripening. Sci Rep 7:16674
Wang TQ, Chen T, Yan HF, Wang Y (2018) TCM treatment of anemopyretic cold rule analysis. J Tianjin Univ Tradit Chin Med 37:113–117
Wang Y, Pang YL, Chen K, Zhai LY, Shen CC, Wang S, Xu JL (2020) Genetic bases of source-, sink-, and yield-related traits revealed by genome-wide association study in Xian rice. Crop J 8:119–131
Wei Q, Fu W, Wang Y, Qin X, Wang J, Li J, Lou Q, Chen J (2016) Rapid identification of fruit length loci in cucumber (Cucumis sativus L.) using next-generation sequencing (NGS)-based QTL analysis. Sci Rep 6:27496
Yan R, Yang YJ, Liu HW, Li XN, Guo BL (2016) Effect of earlier period harvest on content of forsythoside A and phillyrin of forsythiae fructus. Mod Chin Med 18:579–582
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82
Yang L, Li D, Li Y, Gu X, Huang S, Garcia-Mas J, Weng Y (2013) A 1681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol 13:53
Zhang XX, Guan ZR, Li ZL, Liu P, Ma LL, Zhang YC, Pan L, He SJ, Zhang YL, Li P, Ge F, Zou CY, He YC, Gao SB, Pan GT, Shen YO (2020) A combination of linkage mapping and GWAS brings new elements on the genetic basis of yield-related traits in maize across multiple environments. Theor Appl Genet 133:2881–2895
Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44:821–824
Zhu DF, Zhang YP, Chen HZ, Xiang J, Zhang YK (2015) Innovation and practice of high-yield rice cultivation technology in China. Sci Agric Sin 48:3404–3414
Acknowledgements
The authors wish to acknowledge the sampling assistance of Xin Sun.
Funding
This work was supported by the National Natural Science Foundation of China (31770225), Henan Science and Technology Project (202102110077), National Key R&D Program of China (2017FY100705), Foundation of Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences (PCU201903), and Henan Agricultural University Science & Technology Innovation Fund (KJCX2016A2).
Author information
Authors and Affiliations
Contributions
YL conceived the research project, NCP and YL wrote the paper and analyzed the data, QW and HLL collected the data, JEQ, QW, HLL, and YXH revised the paper. All authors are agreed with the content of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Data availability
Sequence data are archived at the National Center for Biotechnology Information (NCBI SRR12897525-12897584).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13258_2021_1186_MOESM1_ESM.docx
Supplementary file1 Fig. S1 All fruit samples of sixty weeping forsythia seedlings. Fig. S2 The optimal clustering at K = 1 were supported based on the cross-validation error rate. Fig. S3 Genotyping of variant loci binding phenotypic data for fruit length. Fig. S4 Genotyping of variant loci binding phenotypic data for fruit width. Fig. S5 Genotyping of variant loci binding phenotypic data for fruit thickness. Fig. S6 Genotyping of variant loci binding phenotypic data for single grain weight (DOCX 921 kb)
13258_2021_1186_MOESM2_ESM.xls
Supplementary file2 Table S1 Phenotypic data of weeping forsythia fruits from sixty weeping forsythia seedlings. Table S2 The variant loci and candidate genes related to fruit length, width, thickness, and single grain weight (XLS 47 kb)
Rights and permissions
About this article
Cite this article
Li, Y., Wu, Q., Liu, HL. et al. Identification of yield-related genes through genome-wide association: case study of weeping forsythia, an emerging medicinal crop. Genes Genom 44, 145–154 (2022). https://doi.org/10.1007/s13258-021-01186-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-021-01186-w