Abstract
Background
As the transformation process can induce mutations in host plants, molecular characterization of the associated genomic changes is important not only for practical food safety but also for understanding the fundamental theories of genome evolution.
Objectives
To investigate a population-scale comparative study of the genome-wide spectrum of sequence variants in the transgenic genome with the variations present in 3000 rice varieties.
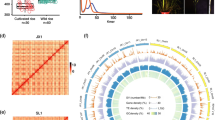
Results
On average, we identified 19,273 SNPs (including Indels) per transgenic line in which 10,729 SNPs were at the identical locations in the three transgenic rice plants. We found that these variations were predominantly present in specific regions in chromosomes 8 and 10. Majority (88%) of the identified variations were detected at the same genomic locations as those in natural rice population, implying that the transgenic induced mutations had a tendency to be common alleles.
Conclusion
Genomic variations in transgenic rice plants frequently occurred at the same sites as the major alleles found in the natural rice population, which implies that the sequence variations occur within the limits of a biological system to ensure survival.




Similar content being viewed by others
Data availability
The datasets supporting the results of this article are available in the NCBI Sequence Read Archive repository [SRX2762614, SRX2762615, SRX2762616, SRX2762617, SRX2762618, and SRX2762619]. The SNP and Indel datasets of the 3K rice genomes are available in a public repository under the link: http://snp-seek.irri.org/_download.zul (Mansueto et al. 2017).
Abbreviations
- SNP:
-
Single nucleotide polymorphism
- Indel:
-
Insertion and deletion
- Mb:
-
Mega base pair
- 3K RGP:
-
3000 rice genomes project
- GM:
-
Genetically modified
- T-DNA:
-
Transfer DNA
- Kb:
-
Kilo base pair
- 2, 4-D:
-
2, 4-Dichlorophenoxyacetic acid
- DSB:
-
Double strand breaks
- BWA:
-
Burrow Wheeler Aligner
- GATK:
-
Genome analysis toolkit
- SEA:
-
Singular enrichment analysis.
References
Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes RR, Ulat VJ, Chebotarov D, Zhang G, Li Z et al (2015) SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res 43(Database issue):D1023–D1027
Amos W (2010) Even small SNP clusters are non-randomly distributed: is this evidence of mutational non-independence? Proc Biol Sci 277:1443–1449
Anderson JE, Michno JM, Kono TJ, Stec AO, Campbell BW, Curtin SJ, Stupar RM (2016) Genomic variation and DNA repair associated with soybean transgenesis: a comparison to cultivars and mutagenized plants. BMC Biotechnol 16:41. https://doi.org/10.1186/s12896-016-0271-z
Arbeithuber B, Betancourt AJ, Ebner T, Tiemann-Boege I (2015) Crossovers are associated with mutation and biased gene conversion at recombination hotspots. Proc Natl Acad Sci USA 112:2109–2114
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2):80–92
Gelvin SB (2010) Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu Rev Phytopathol 48:45–68
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Goodman RM, Hauptli H, Crossway A, Knauf VC (1987) Gene transfer in crop improvement. Science 236:48–54
Gross BL, Zhao Z (2014) Archaeological and genetic insights into the origins of domesticated rice. Proc Natl Acad Sci USA 111:6190–6197
Guttikonda SK, Marri P, Mammadov J, Ye L, Soe K, Richey K, Cruse J, Zhuang MB, Gao ZF, Evans C et al (2016) Molecular characterization of transgenic events using next generation sequencing approach. PLoS ONE 11(2):e0149515. https://doi.org/10.1371/journal.pone.0149515
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z et al (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W et al (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jiang C, Mithani A, Gan X, Belfield EJ, Klingler JP, Zhu JK, Ragoussis J, Mott R, Harberd NP (2011) Regenerant Arabidopsis lineages display a distinct genome-wide spectrum of mutations conferring variant phenotypes. Curr Biol 21:1385–1390
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu JZ, Zhou SG et al (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. https://doi.org/10.1186/1939-8433-6-4
Kawakatsu T, Kawahara Y, Itoh T, Takaiwa F (2013) A Whole-genome analysis of a transgenic rice seed-based edible vaccine against cedar pollen allergy. DNA Res 20:623–631
Kessler DA, Taylor MR, Maryanski JH, Flamm EL, Kahi LS (1992) The safety of foods developed by biotechnology. Science 256:1747–1749
König A, Cockburn A, Crevel RWR, Debruyne E, Grafstroem R, Hammerling U, Kimber I, Knudsen I, Kuiper HA, Peijnenburg AA et al (2004) Assessment of the safety of foods derived from genetically modified (GM) crops. Food Chem Toxicol 42:1047–1088
Kovalchuk I, Kovalchuk O, Hohn B (2000) Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J 19:4431–4438
Labra M, Vannini C, Grassi F, Bracale M, Balsemin M, Basso B, Sala F (2004) Genomic stability in Arabidopsis thaliana transgenic plants obtained by floral dip. Theor Appl Genet 109:1512–1518
Lee DK, Park SH, Seong SY, Kim YS, Jung H, Do Choi Y, Kim JK (2016) Production of insect-resistant transgenic rice plants for use in practical agriculture. Plant Biotechnol Rep 10:391–401
Lercher MJ, Hurst LD (2002) Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet 18:337–340
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li G, Chern M, Jain R, Martin JA, Schackwitz WS, Jiang L, Vega-Sanchez ME, Lipzen AM, Barry KW, Schmutz J et al (2016) Genome-wide sequencing of 41 Rice (Oryza sativa L.) mutated lines reveals diverse mutations induced by fast-neutron irradiation. Mol Plant 9:1078–1081
Li R, Quan S, Yan X, Biswas S, Zhang D, Shi J (2017) Molecular characterization of genetically-modified crops: challenges and strategies. Biotechnol Adv 35:302–309
Mansueto L, Fuentes RR, Borja FN, Detras J, Abriol-Santos JM, Chebotarov D, Sanciangco M, Palis K, Copetti D, Poliakov A et al (2017) Rice SNP-seek database update: new SNPs, indels, and queries. Nucleic Acids Res 45(D1):D1075–D1081. https://doi.org/10.1093/nar/gkw1135
McCouch SR, Wright MH, Tung CW, Maron LG, McNally KL, Fitzgerald M, Singh N, DeClerck G, Agosto-Perez F, Korniliev P et al (2016) Open access resources for genome-wide association mapping in rice. Nat Commun 7:10532
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M et al (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE et al (2009) Genome wide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci USA 106:12273–12278
Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL et al (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452:991–996
Miyao A, Nakagome M, Ohnuma T, Yamagata H, Kanamori H, Katayose Y, Takahashi A, Matsumoto T, Hirochika H (2012) Molecular spectrum of somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol 53:256–264
Molina J, Sikora M, Garud N, Flowers JM, Rubinstein S, Reynolds A, Huang P, Jackson S, Schaal BA, Bustamante CD et al (2011) Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci USA 108:8351–8356
Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94
Park D, Park SH, Ban YW, Kim YS, Park KC, Kim NS, Kim JK, Choi IY (2017) A bioinformatics approach for identifying transgene insertion sites using whole genome sequencing data. BMC Biotechnol 17:67. https://doi.org/10.1186/s12896-017-0386-x
Rattray A, Santoyo G, Shafer B, Strathern JN (2015) Elevated mutation rate during meiosis in Saccharomyces cerevisiae. PLoS Genet 11(1):e1004910. https://doi.org/10.1371/journal.pgen.1004910
Ross-Ibarra J, Morrell PL, Gaut BS (2007) Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci USA 104(Suppl 1):8641–8648
Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang CC, Iwamoto M, Abe T et al (2013) Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol 54:e6
Shee C, Gibson JL, Rosenberg SM (2012) Two mechanisms produce mutation hotspots at DNA breaks in Escherichia coli. Cell Rep 2(4):714–721. https://doi.org/10.1093/pcp/pcs183
Sparrow PA (2010) GM risk assessment. Mol Biotechnol 44:267–275
Sun C, Hu Z, Zheng T, Lu K, Zhao Y, Wang W, Shi J, Wang C, Lu J, Zhang D et al (2017) RPAN: rice pan-genome browser for approximately 3000 rice genomes. Nucleic Acids Res 45:597–605
The 3,000 rice genomes project (2014) The 3,000 rice genomes project. GigaScience 3:7. https://doi.org/10.1186/2047-217X-3-7
Wagner A (2000) Robustness against mutations in genetic networks of yeast. Nat Genet 24:355–361
Weber N, Halpin C, Hannah LC, Jez JM, Kough J, Parrott W (2012) Crop genome plasticity and its relevance to food and feed safety of genetically engineered breeding stacks. Plant Physiol 160:1842–1853
Wei FJ, Kuang LY, Oung HM, Cheng SY, Wu HP, Huang LT, Tseng YT, Chiou WY, Hsieh-Feng V, Chung CH et al (2016) Somaclonal variation does not preclude the use of rice transformants for genetic screening. Plant J 85:648–659
Wolfe KH, Sharp PM, Li WH (1989) Mutation rates differ among regions of the mammalian genome. Nature 337:283–285
Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L et al (2011) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30:105–111
Yamamoto T, Nagasaki H, Yonemaru J, Ebana K, Nakajima M, Shibaya T, Yano M (2010) Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics 11:267. https://doi.org/10.1186/1471-2164-11-267
Zhang D, Wang Z, Wang N, Gao Y, Liu Y, Wu Y, Bai Y, Zhang Z, Lin X, Dong Y et al (2014) Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PLoS ONE 9(5):e96879. https://doi.org/10.1371/journal.pone.0096879
Funding
This work was supported by a grant from the Next-Generation BioGreen21 Program (PJ013658022018 to YSK), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
JKK and IYC conceived and designed the project. DP was a major contributor to the data analysis and drafted the manuscript. SHP and YSK constructed the transgenic plants and prepared sample materials. BSC analyzed the data. NSK analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, D., Park, SH., Kim, Y.S. et al. NGS sequencing reveals that many of the genetic variations in transgenic rice plants match the variations found in natural rice population. Genes Genom 41, 213–222 (2019). https://doi.org/10.1007/s13258-018-0754-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-018-0754-5