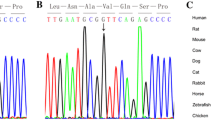
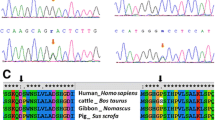
Abstract
Congenital heart disorders (CHDs) are common, estimated to affect 8 out of every 1000 live births, and they are associated with high mortality and morbidity. It is still unknown what causes CHDs. In the pathophysiology of CHD, environmental and inherited factors are both involved. The zinc-finger transcription factor GATA4 is crucial for the early stages of heart morphogenesis. Early expression of GATA4 in the pro-cardiogenic splanchnic mesoderm is necessary for the creation of pre-epicardium during cardiac development. Additionally, it controls the expression of the Wnt 11 gene, which controls early cardiogenesis. Congenital heart disorders are thought to be brought on by GATA4 mutations. The role of GATA4 mutations in the occurrence of congenital cardiac disorders in our environment is not well understood. Case–control GATA4 mutation analysis was performed to shed light on the etiology of sporadic non-syndromic congenital cardiac disorders. Using high-resolution melting assays and sequencing analysis, we have screened exons 7 and 2 of GATA4, which are regarded as molecular hot spots in patients with sporadic congenital heart diseases. In addition, we sequenced GATA4 exon 7, which comprises crucial regulatory components involved in polyadenylation, nuclear transport, translation, and messenger RNA degradation. We have taken into account non-inherited risk factors that are also connected to the pathophysiology of congenital heart disorders, such as folic acid, alcohol, gestational age, birth weight, and maternal age. We have identified missense insertion, silent substitution, and missense substitution of the GATA4 gene in phenotypes, namely; tetralogy of Fallot, ventricular septal defects, and atrial septal defects.

















Similar content being viewed by others
Data availability and materials
Patient data and participant blood samples were obtained at JKCI, and all the references in this manuscript were cited from PubMed and Google Scholar. We used Endnote software version 8 for data management and referencing.
Abbreviations
- ASD:
-
Atrial septal defect
- VSD:
-
Ventral septal defects
- AVSD:
-
Atrial ventral septal defects
- DORV:
-
Double outlet right ventricle
- TGV:
-
Transposition of greater vessels
- TOF:
-
Tetralogy of fallot
- JKCI:
-
Jakaya Kikwete Cardiac Institute
- CHDs:
-
Congenital heart diseases
- DNA:
-
Deoxyribonucleic Acid
- DBS:
-
Dry blood sample
- GATA4:
-
GATA 4 binding protein gene
- MUHAS:
-
Muhimbili University of Health and Allied Sciences
- HRMA:
-
High resolution melting assays
- PAH:
-
Pulmonary arterial hypertension
- PCR:
-
Polymerase chain reaction
- aOR:
-
Adjusted odd ratios
References
Abbasi S, Mohsen-Pour N, Naderi N, Rahimi S, Maleki M, et al. In silico analysis of GATA4 variants demonstrates main contribution to congenital heart disease. J Cardiovasc Thorac Res. 2021;13(4):4.
Afouda BA, Martin J, Liu F, Ciau-Uitz A, Patient R, et al. GATA transcription factors integrate Wnt signalling during heart development. Development. 2008;135(19):3185–90.
Anderson RH, Wenink AC. Thoughts on concepts of development of the heart in relation to the morphology of congenital malformations. Experientia. 1988;44(11–12):951–60.
Bakker MK, Bergman JEH, Krikov S, Amar E, Cocchi G, et al. Prenatal diagnosis and prevalence of critical congenital heart defects: an international retrospective cohort study. BMJ Open. 2019;9(7):e028139.
Bu H, Sun G, Zhu Y, Yang Y, Tan Z, et al. The M310T mutation in the GATA4 gene is a novel pathogenic target of the familial atrial septal defect. BMC Cardiovasc Disord. 2021;21(1):12.
Carmichael SL, Shaw GM, Yang W, Lammer EJ. Maternal periconceptional alcohol consumption and risk for conotruncal heart defects. Birth Defects Res A Clin Mol Teratol. 2003;67(10):875–8.
Chen J, Qi B, Zhao J, Liu W, Duan R, et al. A novel mutation of GATA4 (K300T) associated with familial atrial septal defect. Gene. 2016;575(2 Pt 2):473–7.
Chen MW, Pang YS, Guo Y, Liu BL, Shen J, et al. Association between GATA-4 mutations and congenital cardiac septal defects in Han Chinese patients. Zhonghua Xin Xue Guan Bing Za Zhi. 2009;37(5):409–12.
Chen MW, Pang YS, Guo Y, Pan JH, Liu BL, et al. GATA4 mutations in Chinese patients with congenital cardiac septal defects. Pediatr Cardiol. 2010;31(1):85–9.
Chen Y, Han ZQ, Yan WD, Tang CZ, Xie JY, et al. A novel mutation in GATA4 gene associated with dominant inherited familial atrial septal defect. J Thorac Cardiovasc Surg. 2010;140(3):684–7.
Chen Y, Mao J, Sun Y, Zhang Q, Cheng HB, et al. A novel mutation of GATA4 in a familial atrial septal defect. Clin Chim Acta. 2010;411(21–22):1741–5.
Czeizel AE, Vereczkey A, Szabó I. Folic acid in pregnant women associated with reduced prevalence of severe congenital heart defects in their children: a national population-based case-control study. Eur J Obstet Gynecol Reprod Biol. 2015;193:34–9.
Yan Y, Wu Q, Zhang L, Wang X, Dan S, Deng D, Sun L, Yao L, Ma Y, Wang L. Detection of submicroscopic chromosomal aberrations by array-based comparative genomic hybridization in fetuses with congenital heart disease. Ultrasound Obstet Gynecol. 2014;43(4):404–12.
Durán RMP, Brotons DA, Argüelles IZ, Concepción PM. Advances in pediatric cardiology and congenital heart diseases. Revista Espanola Cardiol. 2008;61(Suppl 1):15–26.
Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol. 2008;85(1):50–8.
Fan D, Pang S, Chen J, Shan J, Cheng Q, et al. Identification and functional study of GATA4 gene regulatory variants in atrial septal defects. BMC Cardiovasc Disord. 2021;21(1):321.
Fox S, Filichkin S, Mockler TC. Applications of ultra-high-throughput sequencing. Methods Mol Biol. 2009;553:79–108.
Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424(6947):443–7.
Gelb BD. Genetic basis of congenital heart disease. Curr Opin Cardiol. 2004;19(2):110–5.
Geng J, Picker J, Zheng Z, Zhang X, Wang J, et al. Chromosome microarray testing for patients with congenital heart defects reveals novel disease causing loci and high diagnostic yield. BMC Genomics. 2014;15:1127.
Gibbs RA. The human genome project changed everything. Nat Rev Genet. 2020;21(10):575–6.
Glessner JT, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115(10):884–96.
Zimmerman MS, Smith AGC, Sable CA, Echko MM, Wilner LB, Olsen HE, Atalay HT, Awasthi A, Bhutta ZA, Boucher JL, Castro F. Global, regional, and national burden of congenital heart disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Child Adolesc Health. 2020;4(3):185–200.
Goetzinger KR, Shanks AL, Odibo AO, Macones GA, Cahill AG. Advanced maternal age and the risk of major congenital anomalies. Am J Perinatol. 2017;34(3):217–22.
Granados-Riveron JT, Pope M, Bu’lock FA, Thornborough C, Eason J, et al. Combined mutation screening of NKX2-5, GATA4, and TBX5 in congenital heart disease: multiple heterozygosity and novel mutations. Congenit Heart Dis. 2012;7(2):151–9.
Grépin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development. 1997;124(12):2387–95.
Grewal J, Carmichael SL, Ma C, Lammer EJ, Shaw GM. Maternal periconceptional smoking and alcohol consumption and risk for select congenital anomalies. Birth Defects Res A Clin Mol Teratol. 2008;82(7):519–26.
Han H, Chen Y, Liu G, Han Z, Zhao Z, et al. GATA4 transgenic mice as an in vivo model of congenital heart disease. Int J Mol Med. 2015;35(6):1545–53.
Jiang Y, Drysdale TA, Evans T. A role for GATA-4/5/6 in the regulation of Nkx2.5 expression with implications for patterning of the precardiac field. Dev Biol. 1999;216(1):57–71.
Jiang Y, Tarzami S, Burch JB, Evans T. Common role for each of the cGATA-4/5/6 genes in the regulation of cardiac morphogenesis. Dev Genet. 1998;22(3):263–77.
Kang H. Sample size determination and power analysis using the G*Power software. J Educ Eval Health Prof. 2021;18:17.
Khatami M, Ghorbani S, Adriani MR, Bahaloo S, Naeini MA, Heidari MM, Hadadzadeh M. Novel point mutations in 3′-untranslated region of GATA4 gene are associated with sporadic non-syndromic atrial and ventricular septal defects. Current Medical Science. 2022;42(1):129–43.
Kramer HH, Trampisch HJ, Rammos S, Giese A. Birth weight of children with congenital heart disease. Eur J Pediatr. 1990;149(11):752–7.
Leirgul E, Brodwall K, Greve G, Vollset SE, Holmstrøm H, et al. Maternal diabetes, birth weight, and neonatal risk of congenital heart defects in Norway, 1994–2009. Obstet Gynecol. 2016;128(5):1116–25.
Liu X, Liu G, Wang P, Huang Y, Liu E, et al. Prevalence of congenital heart disease and its related risk indicators among 90,796 Chinese infants aged less than 6 months in Tianjin. Int J Epidemiol. 2015;44(3):884–93.
Liu XY, Wang J, Zheng JH, Bai K, Liu ZM, et al. Involvement of a novel GATA4 mutation in atrial septal defects. Int J Mol Med. 2011;28(1):17–23.
Liu XY, Yang YQ, Ma J, Lin XP, Zheng JH, et al. Novel GATA4 mutations identified in patients with congenital atrial septal defects. Zhonghua Xin Xue Guan Bing Za Zhi. 2010;38(8):724–7.
Löser H, Pfefferkorn JR, Themann H. Alcohol in pregnancy and fetal heart damage. Klin Padiatr. 1992;204(5):335–9.
Luca AC, Holoc AS, Iordache C. Congenital heart malformations in newborn babies with low birth weight. Rev Med Chir Soc Med Nat Iasi. 2015;119(2):353–60.
Mangili G, Garzoli E, Sadou Y. Feeding dysfunctions and failure to thrive in neonates with congenital heart diseases. Pediatr Med Chir. 2018. https://doi.org/10.4081/pmc.2018.196.
Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402.
Mattapally S, Nizamuddin S, Murthy KS, Thangaraj K, Banerjee SK. c. 620C> T mutation in GATA4 is associated with congenital heart disease in South India. BMC Med Genet. 2015;16(1):1–12.
McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–77.
McElhinney DB. Recent progress in the understanding and management of postoperative right ventricular outflow tract dysfunction in patients with congenital heart disease. Circulation. 2012;125(16):e595–9.
Medoff-Cooper B, Naim M, Torowicz D, Mott A. Feeding, growth, and nutrition in children with congenitally malformed hearts. Cardiol Young. 2010;20(Suppl 3):149–53.
Medoff-Cooper B, Ravishankar C. Nutrition and growth in congenital heart disease: a challenge in children. Curr Opin Cardiol. 2013;28(2):122–9.
Misra C, Sachan N, McNally CR, Koenig SN, Nichols HA, et al. Congenital heart disease-causing gata4 mutation displays functional deficits in vivo. PLoS Genet. 2012;8(5):e1002690.
Mohan RA, van Engelen K, Stefanovic S, Barnett P, Ilgun A, et al. A mutation in the Kozak sequence of GATA4 hampers translation in a family with atrial septal defects. Am J Med Genet A. 2014;164(11):2732–8.
Montgomery JL, Sanford LN, Wittwer CT. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn. 2010;10(2):219–40.
Naghavi-Behzad M, Alizadeh M, Azami S, Foroughifar S, Ghasempour-Dabbaghi K, et al. Risk factors of congenital heart diseases: a case-control study in Northwest Iran. J Cardiovasc Thorac Res. 2013;5(1):5–9.
Nees SN, Chung WK. The genetics of isolated congenital heart disease. Am J Med Genet C Semin Med Genet. 2020;184(1):97–106.
Nemer G, Fadlalah F, Usta J, Nemer M, Dbaibo G, et al. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Human Mutat. 2006;27(3):293–4. https://doi.org/10.1002/humu.9410.
Nora JJ. Multifactorial inheritance hypothesis for the etiology of congenital heart diseases. Circulation. 1968;38(3):604–17.
Obeid R, Holzgreve W, Pietrzik K. Folate supplementation for prevention of congenital heart defects and low birth weight: an update. Cardiovasc Diagn Ther. 2019;9(Suppl 2):S424-s433.
Oberhuber RD, Huemer S, Mair R, Sames-Dolzer E, Kreuzer M, et al. Cognitive development of school-age hypoplastic left heart syndrome survivors: a single center study. Pediatr Cardiol. 2017;38(6):1089–96.
Orjuela Quintero DC, Núñez F, Caicedo V, Pachón S, Salazar SM. Mutations in the GATA4 gen in patients with non-syndromic congenital heart disease. Invest Clin. 2014;55(3):207–16.
Pascall E, Tulloh RM. Pulmonary hypertension in congenital heart disease. Future Cardiol. 2018;14(4):343–53.
Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, et al. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p231 and congenital heart disease. Am J Med Genet. 1999;83(3):201–6. https://doi.org/10.1002/(SICI)1096-8628(19990319)83:3%3c201::AID-AJMG11%3e3.0.CO;2-V.
Pei L, Kang Y, Zhao Y, Yan H. Prevalence and risk factors of congenital heart defects among live births: a population-based cross-sectional survey in Shaanxi province, Northwestern China. BMC Pediatr. 2017;17(1):18.
Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho R. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63(2):196–207.
Pulignani S, Vecoli C, Sabina S, Foffa I, Ait-Ali L, et al. 3’UTR SNPs and haplotypes in the GATA4 gene contribute to the genetic risk of congenital heart disease. Rev Esp Cardiol. 2016;69(8):760–5.
Qu Y, Lin S, Zhuang J, Bloom MS, Smith M, et al. First-trimester maternal folic acid supplementation reduced risks of severe and most congenital heart diseases in offspring: a large case-control study. J Am Heart Assoc. 2020;9(13):e015652.
Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol. 2007;43(6):677–85.
Raphael DM, Roos L, Myovela V, McHomvu E, Namamba J, et al. Heart diseases and echocardiography in rural Tanzania: occurrence, characteristics, and etiologies of underappreciated cardiac pathologies. PLoS ONE. 2018;13(12):e0208931.
Reamon-Buettner SM, Cho S-H, Borlak J. Mutations in the 3’-untranslated region of GATA4 as molecular hotspots for congenital heart disease (CHD). BMC Med Genet. 2007;8(1):38.
Rood Jennifer E, Regev A. The legacy of the human genome project. Science. 2021;373(6562):1442–3.
Sarajuuri A, Jokinen E, Puosi R, Eronen M, Mildh L, et al. Neurodevelopmental and neuroradiologic outcomes in patients with univentricular heart aged 5 to 7 years: related risk factor analysis. J Thorac Cardiovasc Surg. 2007;133(6):1524–32.
Schluterman MK, Krysiak AE, Kathiriya IS, Abate N, Chandalia M, et al. Screening and biochemical analysis of GATA4 sequence variations identified in patients with congenital heart disease. Am J Med Genet Part A. 2007;143A(8):817–23. https://doi.org/10.1002/ajmg.a.31652.
Shabana NA, Shahid SU, Irfan U. Genetic contribution to congenital heart disease (CHD). Pediatr Cardiol. 2020;41(1):12–23.
Soheili F, Jalili Z, Rahbar M, Khatooni Z, Mashayekhi A, et al. Novel mutation of GATA4 gene in Kurdish population of Iran with nonsyndromic congenital heart septals defects. Congenit Heart Dis. 2018;13(2):295–304.
Tang ZH, Xia L, Chang W, Li H, Shen F, et al. Two novel missense mutations of GATA4 gene in Chinese patients with sporadic congenital heart defects. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23(2):134–7.
Tanner K, Sabrine N, Wren C. Cardiovascular malformations among preterm infants. Pediatrics. 2005;116(6):e833-838.
Tantchou Tchoumi JC, Butera G, Giamberti A, Ambassa JC, Sadeu JC. Occurrence and pattern of congenital heart diseases in a rural area of sub-Saharan Africa : cardiovascular topics. Cardiovasc J Afr. 2011;22(2):63–6.
Taylor CF. Mutation scanning using high-resolution melting. Biochem Soc Trans. 2009;37(Pt 2):433–7.
Thomas M, Olusoji O, Awolola N. Spectrum of congenital heart diseases in an African population: a necropsy study. World J Cardiovasc Dis. 2013;03(01):34–9.
Välimäki MJ, Leigh RS, Kinnunen SM, March AR, de Sande AH, et al. GATA-targeted compounds modulate cardiac subtype cell differentiation in dual reporter stem cell line. Stem Cell Res Ther. 2021;12(1):190.
Välimäki MJ, Ruskoaho HJ. Targeting GATA4 for cardiac repair. IUBMB Life. 2020;72(1):68–79.
van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–7.
Vossen RH, Aten E, Roos A, den Dunnen JT. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat. 2009;30(6):860–6.
Wang E, Sun S, Qiao B, Duan W, Huang G, et al. Identification of functional mutations in GATA4 in patients with congenital heart disease. PLoS ONE. 2013;8(4):e62138.
Wang J, Hu DY, Li XM, Xin YF, Zhou H, et al. Novel GATA4 mutations identified in patients with congenital heart disease. Zhonghua Yi Xue Za Zhi. 2010;90(10):667–71.
Wang J, Li XM, Xin YF, Wang LJ, Xu WJ, et al. Genetic screening for novel GATA4 mutations associated with congenital atrial septal defect. Zhonghua Xin Xue Guan Bing Za Zhi. 2010;38(5):429–34.
Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci USA. 2004;101(34):12573–8.
Whitcomb J, Gharibeh L, Nemer M. From embryogenesis to adulthood: critical role for GATA factors in heart development and function. IUBMB Life. 2020;72(1):53–67.
Xiong F, Li Q, Zhang C, Chen Y, Li P, et al. Analyses of GATA4, NKX2.5, and TFAP2B genes in subjects from southern China with sporadic congenital heart disease. Cardiovasc Pathol. 2013;22(2):141–5.
Yang J, Qiu H, Qu P, Zhang R, Zeng L, et al. Prenatal alcohol exposure and congenital heart defects: a meta-analysis. PLoS ONE. 2015;10(6):e0130681.
Yang Y-Q, Wang M-Y, Zhang X-L, Tan H-W, Shi H-F, et al. GATA4 loss-of-function mutations in familial atrial fibrillation. Clin Chim Acta. 2011;412(19):1825–30.
Yang YQ, Gharibeh L, Li RG, Xin YF, Wang J, et al. GATA4 loss-of-function mutations underlie familial tetralogy of fallot. Hum Mutat. 2013;34(12):1662–71.
Zhang L, Tümer Z, Jacobsen JR, Andersen PS, Tommerup N, et al. Screening of 99 Danish patients with congenital heart disease for GATA4 mutations. Genet Test. 2006;10(4):277–80.
Zhang W, Li X, Shen A, Jiao W, Guan X, et al. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51(6):527–35.
Zhang WM, Li XF, Ma ZY, Zhang J, Zhou SH, et al. GATA4 and NKX2.5 gene analysis in Chinese Uygur patients with congenital heart disease. Chin Med J (Engl). 2009;122(4):416–9.
Zhang X, Wang J, Wang B, Chen S, Fu Q, et al. A Novel Missense Mutation of GATA4 in a Chinese Family with Congenital Heart Disease. PLoS ONE. 2016;11(7):e0158904.
Zuechner A, Mhada T, Majani NG, Sharau GG, Mahalu W, et al. Spectrum of heart diseases in children presenting to a paediatric cardiac echocardiography clinic in the Lake Zone of Tanzania: a 7 years overview. BMC Cardiovasc Disord. 2019;19(1):291.
Zühlke L, Mirabel M, Marijon E. Congenital heart disease and rheumatic heart disease in Africa: recent advances and current priorities. Heart. 2013;99(21):1554.
Acknowledgements
We thank the University of Shandong, the University of Muhimbili, and the Jakaya Kikwete Cardiac Institute for providing the facilities and guidance required for effective writing and submitting the article for publication.
Funding
The corresponding author received funds for writing this manuscript from the Chinese Government Scholarship (CSC).
Author information
Authors and Affiliations
Contributions
All the authors participated in the whole process of writing the manuscript. Emmanuel Suluba (ES) and Weinjia Liang; concept, drafting the manuscript, and manuscript writing; James Masaganya (JM); literature review and manuscript writing; Mwinyi Masala (MM); molecular methods review; Naizihijwa Majani (NM); clinical method review; Erasto Mbugi (EM) and Teddy Mselle (TM); manuscript collections and supervision; Benezeth M. Mutayoba (BM); provide technical support, supervision on molecular techniques, Liu Shuwei (LS), provide guidelines, drafting, manuscript corrections and supervision。
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Consent for publication
All authors consented to publish.
Ethical approval and consent to participate
Ethical clearance for conducting this study was obtained from the Muhimbili University Institutional Review Board (MUHAS-IRB) (Ref.No.DA.282/298/01.C/MUHAS-REC-06–2021-666). Permission to collect data was obtained from the Director of the Jakaya Kikwete Cardiac Institute research and publication committee (Ref: AB.123/307/01E/16). Participants were informed about the aim of the study and their rights, including the right to withdraw from the study at any stage without losing any benefit from the study. We asked the participants to provide consent to allow them and their children to participate in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Corresponding Editor: Ansuman chattopadhyay: Reviewers: Arindam Bandyopadhyay,Pritha Bhattacharjee.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Suluba, E., Masaganya, J., Liang, W. et al. High throughput mutation screening of cardiac transcription factor GATA4 among Tanzania children with congenital heart diseases. Nucleus 66, 11–30 (2023). https://doi.org/10.1007/s13237-022-00414-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-022-00414-2