Abstract
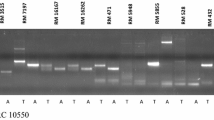
Pod borer, caused by Helicoverpa armigera (Hubner), is an important agronomic pest of pigeonpea. Genetic analysis for resistance to H. armigera in an interspecific F2 population and a set of F3 families both derived from a cross between Cajanus cajan cv. ICP-26 (Susceptible) and C. scarabaeoides acc. ICPW-94 (Resistant) revealed that the resistance is controlled by a dominant allele at a single locus. Pod borer resistance is associated with non glandular short trichomes, which is also governed by a dominant allele at a single locus. The resistance locus ( PPB1 ) was mapped by linkage analysis with 32 random amplified polymorphic DNA (RAPD) loci, and six inter simple sequence repeat (ISSR) loci segregating among the F2 population. The PPB1 locus was mapped to the linkage group 2 (LG-2) and was flanked by ISSR marker loci UBC8731270 (15.9 cM), and the non glandular short trichome (NGST) locus (12.0 cM) and OPA18565 (26.3 cM). The presence of only few double recombinants between UBC8731270 and NGST (4.31 %) and PPB1 and OPA18565 (10.34 %) in the F2 population indicated that the simultaneous use of both the markers along with morphological trait NGST would be useful in introgression of pod borer resistance gene into C. cajan background and pigeonpea breeding programme.




Similar content being viewed by others
References
Aruna R, Rao DM, Reddy LJ, Upadhyaya HD, Sharma HC. Inheritance of trichomes and resistance to pod borer (Helicoverpa armigera) and their association in interspecific crosses between cultivated pigeonpea (Cajanus cajan) and its wild relative C. scarabaeoides. Euphytica. 2005;145:247–57.
Banu MR, Muthiah AR, Ashok S. Evaluation of pigeonpea (Cajanus cajan L.) genotypes against gram pod borer (Helicoverpa armigera). In: Proceedings of 4th International Food legume Research Conference on Food legumes for Nutritional Security, New Delhi, India; 2005. pp. 317–318.
Dhanasekar P, Dhumal KN, Reddy KS. Identification of RAPD markers linked to plant type gene in pigeonpea. Indian J Biotech. 2010;9:58–63.
Ganapathy KN, Byre GM, Venkatesha SC, Ramachandra R, Gnanesh BN, Girish G. Identification of AFLP markers linked sterility mosaic disease in pigeonpea [Cajanus cajan (L.) Millsp.]. Intl J Integr Biol. 2009;7:145–9.
Gnanesh BN, Bohra A, Sharma M, Byregowda M, Pandey S, Wesley V, et al. Genetic mapping and quantitative trait locus analysis of resistance to sterility mosaic disease in pigeonpea [Cajanus cajan (L.) Millsp.]. Field Crops Res. 2011;123:53–61.
Grover A, Pental D. Breeding objectives and requirements for producing transgenics for major field crops of India. Curr Sci. 2003;84:310–20.
Kole C, Gupta PK. Genome mapping and map based cloning. In: Jain HK, Khrakwal MC, editors. Plant breeding- Mendelian to molecular approaches. New Delhi: Narosa Publising House; 2004. p. 257–99.
Kosambi D. The estimation of map distance from recombination values. Ann Eugenics. 1944;12:172–5.
Kotresh H, Fakrudin B, Punnuri SM, Rajkumar BK, Thudi M, Paramesh H, et al. Identification of two RAPD markers genetically linked to a recessive allele of a Fusarium wilt resistance gene in pigeonpea (Cajanus cajan L. Millsp.). Euphytica. 2006;149:113–20.
Lander ES, Green P, Abrahamson J, Barlow A, Dally MJ, Lincoln SE, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–81.
Michellemore RW, Paran I, Kesseli RV. Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88:9828–32.
Panigrahi J, Mishra RR, Sahu AR, Rath SC, Kole C. Marker-Assisted Breeding (MAB) for Stress resistance in crop plants. In: Rout GR, Das AB, editors. Molecular stress physiology of plants. India: Springer; 2013. p. 387–426.
Saxena KB. Genetic improvement of pigeonpea-a review. Trop Plant Biol. 2008;1:159–78.
Sharma OP, Gopali JB, Yelshetty S, Bambawale OM, Garg DK, Bhosle BB. Pests of pigeonpea and their management, NCIPM, LBS building. New Delhi: IARI; 2010.
Sharma HC, Pampapathy G, Reddy LJ. Wild relatives of pigeonpea as a source of resistance to the pod fly (Melanagromyza obtuse Malloch) and pod wasp (Tanaostigmodes cajaninae La Salle). Genet Resour Crop Evol. 2003;50:817–24.
Sivaramakrishnan S, Seetha K, Rao AN, Singh L. RFLP analysis of Cytoplasmic male-sterile lines of pigeonpea [Cajanus cajan (L.) Millsp.] developed by interspecific crosses. Euphytica. 1997;93:307–12.
Varshney RK, Graner A, Sorrells ME. Genomics assisted breeding for crop improvement. Trends Plant Sci. 2005;10:621–30.
Varshney RK, Penmetsa RV, Dutta S, Kulwal PL, Saxena RK, Datta S, et al. Pigeonpea genomics initiative (PGI): an international effort to improve crop productivity of pigeonpea (Cajanus cajan L.). Mol Breed. 2010;26:393–408.
Verulkar SB, Singh DP, Bhattacharya AK. Inheritance of resistance to pod fly and pod borer in the interspecific cross of pigeonpea. Theor Appl Genet. 1997;95:506–8.
Williams JGK, Kubelik AR, Livak KJ, Rafalaski JA, Tingey SV. DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;13:6531–3.
Acknowledgments
The authors are highly grateful to the Department of Biotechnology, and University Grant Commission, Govt. of India, New Delhi for financial assistance though R& D projects (F. No. BT/PR13468/AGR/02/702/2010 of DBT &F No. 37-331/2009 of UGC) to the corresponding author (JP). The authors also acknowledged the gratefulness to Dr. H. D. Upadhyaya, Principal Scientist, ICRISAT for the germplasm.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mishra, R.R., Sahu, A.R., Rath, S.C. et al. Molecular mapping of locus controlling resistance to Helicoverpa armigera (Hubner) in Cajanus cajan L. (Millspaugh) using interspecific F2 mapping population. Nucleus 56, 91–97 (2013). https://doi.org/10.1007/s13237-013-0086-4
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-013-0086-4