Abstract
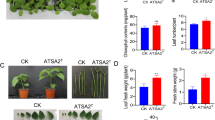
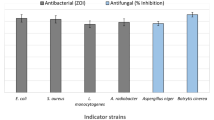
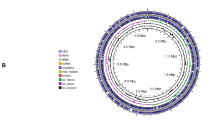
Plant-associated bacteria play an important role in the enhancement of plant growth and productivity. Gluconacetobacter azotocaptans is an exceptional bacterium considering that till today it has been isolated and reported only from Mexico and Canada. It is a plant growth-promoting bacterium and can be used as biofertilizer for different crops and vegetables. The objective of the current study was to evaluate the inoculation effect of Gluconacetobacter azotocaptans DS1, Pseudomonas putida CQ179, Azosprillium zeae N7, Azosprillium brasilense N8, and Azosprillium canadense DS2, on the growth of vegetables including cucumber, sweet pepper, radish, and tomato. All strains increased the vegetables’ growth; however, G. azotocaptans DS1 showed better results as compared to other inoculated and control plants and significantly increased the plant biomass of all vegetables. Therefore, the whole genome sequence of G. azotocaptans DS1 was analyzed to predict genes involved in plant growth promotion, secondary metabolism, antibiotics resistance, and bioremediation of heavy metals. Results of genome analysis revealed that G. azotocaptans DS1 has a circular chromosome with a size of 4.3 Mbp and total 3898 protein-coding sequences. Based on functional analysis, genes for nitrogen fixation, phosphate solubilization, indole acetic acid, phenazine, siderophore production, antibiotic resistance, and bioremediation of heavy metals including copper, zinc, cobalt, and cadmium were identified. Collectively, our findings indicated that G. azotocaptans DS1 can be used as a biofertilizer and biocontrol agent for growth enhancement of different crops and vegetables.



Similar content being viewed by others
References
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Pyshkin AV (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477
Bauer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4):493–496
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Duran HGS, De Los Santos EL, Kim HU, Nave M (2017) antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45(W1):W36–W41
Burnley LE (2000) Heavy metal resistance in the genus Gluconobacter. Thesis, Master of Science in Biology, Faculty of Virginia Tech, Blacksburg, VA, USA
Cardozo VF, Oliveira AG, Nishio EK, Perugini MR, Andrade CG, Silveira WD, Durán N, Andrade G, Kobayashi RK, Nakazato G (2013) Antibacterial activity of extracellular compounds produced by a Pseudomonas strain against methicillin-resistant Staphylococcus aureus (MRSA) strains. Ann Clin Microbiol Antimicrob 12:12
Cavalcante VA, Dobereiner J (1988) A new acid tolerant nitrogen fixing bacterium associated with sugarcane. Plant Soil 108(1988):23–31
Chandra H, Kumari P, Bisht R, Prasad R, Yadav S (2020) Plant growth promoting Pseudomonas aeruginosa from Valeriana wallichii displays antagonistic potential against three phytopathogenic fungi. Mol Biol Rep 47:6015–6026
Chong TM, Yin WF, Chen JW, Mondy S, Grandclément C, Faure D, Dessaux Y, Chan KG (2016) Comprehensive genomic and phenotypic metal resistance profile of Pseudomonas putida strain S13.1.2 isolated from a vineyard soil. AMB Express 6(1):95
Darling AE, Mau B, Perna NT (2010) Progressive Mauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS ONE 5(6):e11147
Duan J, Jiang W, Cheng Z, Heikkila JJ, Glick BR (2013) The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS ONE 8(3):e58640
Eida AA, Bougouffa S, L’Haridon F, Alam I, Weisskopf L, Bajic VB, Saad MM, Hirt H (2020) Genome insights of the plant-growth promoting bacterium Cronobacter muytjensii JZ38 with volatile-mediated antagonistic activity against Phytophthora infestans. Front Microbiol 11:369
El-Sayed M, Helal M (2016) Multiple heavy metal and antibiotic resistance of Acinetobacter baumannii Strain HAF-13 isolated from industrial effluents. Am J Microbiol Res 4:26–36
Eskin N, Vessey K, Tian L (2014) Research progress and perspectives of nitrogen fixing bacterium, Gluconacetobacter diazotrophicus, in monocot plants. Int J Agron 2014:208383
Fuchs G, Boll M, Heider J (2011) Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9(11):803–816
Fuentes-Ramirez LE, Bustillos-Cristales R, Tapia-Hernandez A, Jimenez-Salgado T, Wang ET, Martinez-Romero E, Caballero Mellado J (2001) Novel nitrogen-fixing acetic acid bacteria, Gluconacetobacter johannae sp. nov. and Gluconacetobacter azotocaptans sp. nov., associated with coffee plants. Int J Syst Evol Microbiol 51(Pt 4):1305–1314
Ge X, Zhao Y, Hou W, Zhang W, Chen W, Wang J et al (2013) Complete genome sequence of the industrial strain Gluconobacter oxydans H24. Genome Announc 1(1):e00003-13
Giongo A, Tyler HL, Zipperer UN, Triplett EW (2010) Two genome sequences of the same bacterial strain, Gluconacetobacter diazotrophicus PAL 5, suggest a new standard in genome sequence submission. Stand Gen Sci 2(3):309–317
Glick BR (2010) Using soil bacteria to facilitate phytoremediation. Biotechnol Adv 28(3):367–374
Gonzalez AJ, Larraburu EE, Llorente BE (2015) Azospirillum brasilense increased salt tolerance of jojoba during in vitro rooting. Indian Crop Prod 76:41–48
Guo DJ, Singh RK, Singh P, Li DP, Sharma A, Xing YX, Song XP, Yang LT, Li YR (2020) Complete genome sequence of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front Microbiol 11:580081
Illeghems K, De Vuyst L, Weckx S (2013) Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics 14:526
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2018) High-throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF et al (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:354–357
Kang SM, Asaf S, Khan AL, Lubna KA, Mun BG et al (2020) Complete genome sequence of Pseudomonas psychrotolerans CS51, a plant growth-promoting bacterium, under heavy metal stress conditions. Microorganisms 8:382
Laili NS, Radziah O, Zaharah SS (2017) Isolation and characterization of plant growth promoting rhizobacteria (PGPR) and their effects on growth of strawberry (Fragaria ananassa Duch.). Bangladesh J Bot 46:277–282
Matsutani M, Suzuki H, Yakushi T, Matsushita K (2014) Draft genome sequence of Gluconobacter thailandicus NBRC 3257. Stand Genomic Sci 9(3):614–623
Mehnaz S, Lazarovits G (2006) Inoculation effects of Pseudomonas putida, Gluconacetobacter azotocaptans and Azospirillum lipoferum on corn plant growth under greenhouse conditions. Microb Ecol 51:326–335
Mehnaz S, Lazarovits G (2017) Gluconacetobacter azotocaptans: a plant growth-promoting bacteria. In: Mehnaz S (ed) Rhizotrophs: plant growth promotion to bioremediation microorganisms for sustainability, vol 2. Springer, Singapore, pp 1–14
Mehnaz S, Weselowski B, Lazarovits G (2006) Isolation of Gluconacetobacter azotocaptans from corn rhizosphere. Syst Appl Microbiol 29(6):496–501
Mehnaz S, Weselowski B, Lazarovits G (2007) Azospirillum zeae sp. nov., diazotrophic bacteria isolated from rhizosphere soil of Zea mays. Int J Syst Evol Microbiol 57(12):2805–2809
Mehnaz S, Baig DN, Lazarovits G (2010) Genetic and phenotypic diversity of plant growth promoting rhizobacteria isolated from sugarcane plants growing in Pakistan. J Microbiol Biotechnol 20:1614–1623
Miura H, Mogi T, Ano Y, Migita CT, Matsutani M, Yakushi T et al (2013) Cyanideinsensitive quinol oxidase (CIO) from Gluconobacter oxydans is a unique terminal oxidase subfamily of cytochrome bd. J Biochem 153:535–545
Mogi T, Ano Y, Nakatsuka T, Toyama H, Muroi A, Miyoshi H et al (2009) Biochemical and spectroscopic properties of cyanide-insensitive quinol oxidase from Gluconobacter oxydans. J Biochem 146:263–271
Morley RE (2013) Impact of free-living diazotrophs, Azospirillum lipoferum and Gluconacetobacter azotocaptans, on growth and nitrogen utilization by wheat (Triticum aestivum cv. Lillian). Dissertation, University of Saskatchewan
Mukhtar S, Shahid I, Mehnaz S, Malik KA (2017) Assessment of two carrier materials for phosphate solubilizing biofertilizers and their effect on growth of wheat (Triticum aestivum). Microbiol Res 205:107–117
Mukhtar S, Ahmad S, Bashir A, Mirza MS, Mehnaz S, Malik KA (2019) Identification of plasmid encoded osmoregulatory genes from halophilic bacteria isolated from the rhizosphere of halophytes. Microbiol Res 228:26307
Mukhtar S, Zareen M, Khaliq Z, Mehnaz S, Malik KA (2020) Phylogenetic analysis of halophyte-associated rhizobacteria and effect of halotolerant and halophilic phosphate-solubilizing biofertilizers on maize growth under salinity stress conditions. Appl Microbiol 128:556–573
Muthukumarasamy R, Revathi G, Seshadri S, Simhan CL (2002) Gluconacetobacter diazotrophicus (syn. Acetobacter diazotrophicus), a promising diazotrophic endophyte in tropics. Curr Sci 83(2):137–145
Muthukumarasamy R, Cleenwerck I, Revathi G et al (2005) Natural association of Gluconacetobacter diazotrophicus and diazotrophic Acetobacter peroxydans with wetland rice. Syst Appl Microbiol 28:277–286
Peters B, Mientus M, Kostner D, Junker A, Liebl W, Ehrenreich A (2013) Characterization of membrane bound dehydrogenases from Gluconobacter oxydans 621H via whole-cell activity assays using multideletion strains. Appl Microbiol Biotechnol 97:6397–6412
Plata G, Henry CS, Vitkup D (2015) Long-term phenotypic evolution of bacteria. Nature 517(7534):369–372
Shahid I, Rizwan M, Baig DN, Saleem RS, Malik KA, Mehnaz S (2017) Secondary metabolites production and plant growth promotion by Pseudomonas chlororaphis subsp. aurantiaca strains isolated from cotton, cactus and para grass. J Microbiol Biotechnol 27:480–491
Singh RK, Singh P, Li HB, Guo DJ, Song QQ, Yang LT et al (2020) Plant-PGPR interaction study of plant growth-promoting diazotrophs Kosakonia radicincitans BA1 and Stenotrophomonas maltophilia COA2 to enhance growth and stress-related gene expression in Saccharum spp. J Plant Interact 15:427–445
Taghavi S, Garafola C, Monchy S (2009) Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl Environ Microbiol 75:748–757
Wang X, Wang C, Li Q, Zhang J, Ji C, Sui J, Liu Z, Song X, Liu X (2018) Isolation and characterization of antagonistic bacteria with the potential for biocontrol of soil-borne wheat diseases. J Appl Microbiol 125(6):1868–1880
Xie H, Pasternak JJ, Glick BR (1996) Isolation and characterization of mutants of the plant growth promoting rhizobacterium Pseudomonas putida GR12-2 that overproduce indoleacetic acid. Curr Microbiol 32(2):67–71
Xu YB, Chen M, Zhang Y, Wang M, Wang Y, Huang QB et al (2014) The phosphotransferase system gene ptsI in the endophytic bacterium Bacillus cereus is required for biofilm formation, colonization, and biocontrol against wheat sharp eyespot. FEMS Microbiol Lett 354:142–152
Yamada Y, Yukphan P, Vu HTL, Muramatsu Y, Ochaikul D, Tanasupawat S, Nakagawa Y (2012) Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J Gen Appl Microbiol 58(5):397–404
Yssel AEJ, Vanderleyden J, Steenackers HP (2017) Repurposing of nucleoside- and nucleobase-derivative drugs as antibiotics and biofilm inhibitors. J Antimicrob Chemother 72:2156–2170
Zanker H, Lurz G, Langridge U, Langridge P, Kreusch D, Schröder J (1994) Octopine and nopaline oxidases from Ti plasmids of Agrobacterium tumefaciens: molecular analysis, relationship, and functional characterization. J Bacteriol 176(15):4511–4517
Acknowledgements
We are highly thankful to the Higher Education Commission, Pakistan (HEC; Project No. 20-3134) and Agriculture and Agri-Food Canada for funding of this research work. We also gratefully acknowledge Dr. Kamran Azeem (Mohammad Ali Jinnah University, Karachi, Pakistan) for his help in sequence analysis and bioinformatics work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mukhtar, S., Farooq, M., Baig, D.N. et al. Whole genome analysis of Gluconacetobacter azotocaptans DS1 and its beneficial effects on plant growth. 3 Biotech 11, 450 (2021). https://doi.org/10.1007/s13205-021-02996-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02996-1