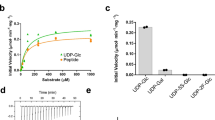
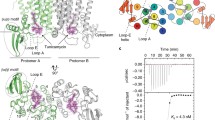
Abstract
Glycosyltransferases (GTs) are widely present in several organisms. These enzymes specifically transfer sugar moieties to a range of substrates. The processes of bacterial glycosylation of the cell wall and their relations with host–pathogen interactions have been studied extensively, yet the majority of mycobacterial GTs involved in the cell wall synthesis remain poorly characterized. Glycopeptidolipids (GPLs) are major class of glycolipids present on the cell wall of various mycobacterial species. They play an important role in drug resistance and host–pathogen interaction virulence. Gtf3 enzyme performs a key step in the biosynthesis of triglycosylated GPLs. Here, we describe a general procedure to achieve expression, purification, and crystallization of recombinant protein Gtf3 from Mycobacterium smegmatis using an E. coli expression system. We reported also a combined bioinformatics and biochemical methods to predict aggregation propensity and improve protein solubilization of recombinant Gtf3. NVoy, a carbohydrate-based polymer reagent, was added to prevent protein aggregation by binding to hydrophobic protein surfaces of Gtf3. Using intrinsic tryptophan fluorescence quenching experiments, we also demonstrated that Gtf3-NVoy enzyme interacted with TDP and UDP nucleotide ligands. This case report proposes useful tools for the study of other glycosyltransferases which are rather difficult to characterize and crystallize.








Similar content being viewed by others
References
Ali ZM, Bakli M, Fontaine A, Bakkali N, Hai VV, Audebert S, Boublik Y, Pagès F, Remoué F, Rogier C (2012) Assessment of Anopheles salivary antigens as individual exposure biomarkers to species-specific malaria vector bites. Malaria J 11(1):439
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42(W1):W252–W258
Billman-Jacobe H (2004) Glycopeptidolipid synthesis in mycobacteria. Curr Sci 111–114
Bitard-Feildel T, Lamiable A, Mornon JP, Callebaut I (2018) Order in disorder as observed by the “hydrophobic cluster analysis” of protein sequences. Proteomics 18(21–22):1800054
Bramucci E, Paiardini A, Bossa F, Pascarella S (2012) PyMod: sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinf 13(4):S2
Brennan PJ, Crick DC (2007) The cell-wall core of Mycobacterium tuberculosis in the context of drug discovery. Curr Top Med Chem 7(5):475–488
Breton C, Šnajdrová L, Jeanneau C, Koča J, Imberty A (2006) Structures and mechanisms of glycosyltransferases. Glycobiology 16(2):29R–37R
Burgess RR (2018) A brief practical review of size exclusion chromatography: rules of thumb, limitations, and troubleshooting. Protein Expr Purif 150:81–85
Chen Y-L, Chen Y-H, Lin Y-C, Tsai K-C, Chiu H-T (2009) Functional characterization and substrate specificity of spinosyn rhamnosyltransferase by in vitro reconstitution of spinosyn biosynthetic enzymes. J Biol Chem 284(11):7352–7363
Deller MC, Kong L, Rupp B (2016) Protein stability: a crystallographer's perspective. Acta Crystallogr Sect F Struct Biol Commun 72(2):72–95
Deshayes C, Laval F, Montrozier H, Daffe M, Etienne G, Reyrat JM (2005) A glycosyltransferase involved in biosynthesis of triglycosylated glycopeptidolipids in Mycobacterium smegmatis: impact on surface properties. J Bacteriol 187(21):7283–7291. https://doi.org/10.1128/JB.187.21.7283-7291.2005
DiMaio F, Leaver-Fay A, Bradley P, Baker D, André I (2011) Modeling symmetric macromolecular structures in Rosetta3. PLoS One 6 (6)
Dolzan M, Johansson K, Roig-Zamboni V, Campanacci V, Tegoni M, Schneider G, Cambillau C (2004) Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function. FEBS Lett 571(1–3):141–146
Gantt RW, Peltier-Pain P, Singh S, Zhou M, Thorson JS (2013) Broadening the scope of glycosyltransferase-catalyzed sugar nucleotide synthesis. Proc Natl Acad Sci USA 110(19):7648–7653. https://doi.org/10.1073/pnas.1220220110
Guild K, Zhang Y, Stacy R, Mundt E, Benbow S, Green A, Myler PJ (2011) Wheat germ cell-free expression system as a pathway to improve protein yield and solubility for the SSGCID pipeline. Acta Crystallogr Sect F Struct Biol Cryst Commun 67(9):1027–1031
Gutiérrez AV, Viljoen A, Ghigo E, Herrmann J-L, Kremer L (2018) Glycopeptidolipids, a double-edged sword of the Mycobacterium abscessus complex. Front Microbiol 9
He C, Liu N, Li F, Jia X, Peng H, Liu Y, Xiao Y (2019) Complex structure of Pseudomonas aeruginosa arginine rhamnosyltransferase EarP with its acceptor elongation factor P. J Bacteriol 201 (13)
Isiorho EA, Jeon B-S, Kim NH, Liu H-w, Keatinge-Clay AT (2014) Structural studies of the spinosyn forosaminyltransferase. SpnP Biochem 53(26):4292–4301
Isiorho EA, Liu H-w, Keatinge-Clay AT (2012) Structural studies of the spinosyn rhamnosyltransferase. SpnG Biochem 51(6):1213–1222
Jeevarajah D, Patterson JH, McConville MJ, Billman-Jacobe H (2002) Modification of glycopeptidolipids by an O-methyltransferase of Mycobacterium smegmatis. Microbiology 148(10):3079–3087. https://doi.org/10.1099/00221287-148-10-3079
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modelling, prediction and analysis. Nat Protoc 10(6):845–858. https://doi.org/10.1038/nprot.2015.053
Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32 (suppl_2):W526-W531
Klammt C, Perrin MH, Maslennikov I, Renault L, Krupa M, Kwiatkowski W, Stahlberg H, Vale W, Choe S (2011) Polymer-based cell-free expression of ligand-binding family B G-protein coupled receptors without detergents. Protein Sci 20(6):1030–1041. https://doi.org/10.1002/pro.636
Kushwaha M, Salis HM (2015) A portable expression resource for engineering cross-species genetic circuits and pathways. Nat Commun 6:7832
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685
Lairson L, Henrissat B, Davies G, Withers S (2008) Glycosyltransferases: structures, functions, and mechanisms. Ann Rev Biochem 77
Lieutaud P, Canard B, Longhi S (2008) MeDor: a metaserver for predicting protein disorder. BMC Genom 9(2):S25
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucl Acids Res 42(D1):D490–D495
Matsui D, Nakano S, Dadashipour M, Asano Y (2017) Rational identification of aggregation hotspots based on secondary structure and amino acid hydrophobicity. Sci Rep 7(1):9558
Miyamoto Y, Mukai T, Nakata N, Maeda Y, Kai M, Naka T, Yano I, Makino M (2006) Identification and characterization of the genes involved in glycosylation pathways of mycobacterial glycopeptidolipid biosynthesis. J Bacteriol 188(1):86–95. https://doi.org/10.1128/JB.188.1.86-95.2006
Mukherjee R, Chatterji D (2005) Evaluation of the role of sigma B in Mycobacterium smegmatis. Biochem Biophys Res Commun 338(2):964–972
Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM (2003) Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc Natl Acad Sci 100(16):9238–9243
Mulichak AM, Losey HC, Walsh CT, Garavito RM (2001) Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure 9(7):547–557
Qasba PK, Ramakrishnan B, Boeggeman E (2005) Substrate-induced conformational changes in glycosyltransferases. Trends Biochem Sci 30(1):53–62
Ren B, Pham TM, Surjadi R, Robinson CP, Le T, Chandry P, Peat TS, McKinstry WJ (2013) Expression, purification, crystallization and preliminary X-ray diffraction analysis of a lactococcal bacteriophage small terminase subunit. Acta Crystallogr Sect F Struct Biol Cryst Commun 69(3):275–279
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172
Schmid J, Heider D, Wendel NJ, Sperl N, Sieber V (2016) Bacterial glycosyltransferases: challenges and opportunities of a highly diverse enzyme class toward tailoring natural products. Front Microbiol 7:182
Schorey JS, Sweet L (2008) The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis. Glycobiology 18(11):832–841
Schuman B, Alfaro JA, Evans SV (2006) Glycosyltransferase structure and function. In: Bioactive Conformation I. Springer, pp 217–257
Sengoku T, Suzuki T, Dohmae N, Watanabe C, Honma T, Hikida Y, Yamaguchi Y, Takahashi H, Yokoyama S, Yanagisawa T (2018) Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP. Nat Chem Biol 14(4):368–374
Sulzenbacher G, Gruez A, Roig-Zamboni V, Spinelli S, Valencia C, Pagot F, Vincentelli R, Bignon C, Salomoni A, Grisel S (2002) A medium-throughput crystallization approach. Acta Crystallogr D Biol Crystallogr 58(12):2109–2115
Thayer KM (2016) Structure prediction and analysis of neuraminidase sequence variants. Biochem Mol Biol Educ 44(4):361–376
Vandermies M, Fickers P (2019) Bioreactor-scale strategies for the production of recombinant protein in the yeast Yarrowia lipolytica. Microorganisms 7(2):40
Veesler D, Blangy S, Siponen M, Vincentelli R, Cambillau C, Sciara G (2009) Production and biophysical characterization of the CorA transporter from Methanosarcina mazei. Anal Biochem 388(1):115–121
Vincentelli R, Canaan S, Campanacci V, Valencia C, Maurin D, Frassinetti F, Scappucini-Calvo L, Bourne Y, Cambillau C, Bignon C (2004) High-throughput automated refolding screening of inclusion bodies. Protein Sci 13(10):2782–2792
Warne NW, Mahler H-C (2018) Challenges in protein product development, vol 38. Springer, New York
Young CL, Britton ZT, Robinson AS (2012) Recombinant protein expression and purification: a comprehensive review of affinity tags and microbial applications. Biotechnol J 7(5):620–634
Zhu F, Erlandsen H, Ding L, Li J, Huang Y, Zhou M, Liang X, Ma J, Wu H (2011) Structural and functional analysis of a new subfamily of glycosyltransferases required for glycosylation of serine-rich streptococcal adhesins. J Biol Chem 286(30):27048–27057
Zhu F, Wu R, Zhang H, Wu H (2013) Structural and biochemical analysis of a bacterial glycosyltransferase. In: Glycosyltransferases. Springer, pp 29–39
Acknowledgments
We would like to thank Dr. Yves Bourne from AFMB laboratory facilities, Dr. Badreddine Douzi and Dr. Renaud Vincentelli for support in protein expression and purification, Dr. Silvia Spinelli for crystallization facilities as well as Stéphanie Blangy, and Dr. David Veesler for support in MALS/UV/refractometry/SEC.
Funding
No funding supported.
Author information
Authors and Affiliations
Contributions
MB and FV conceived and designed the experiments. MB performed the experiments. MB, LK, and FV analyzed the data. MB, LK, NMS, HM, and FV wrote the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors have declared no conflict of interest.
Availability of data and material
All data and materials are available.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bakli, M., Karim, L., Mokhtari-Soulimane, N. et al. Biochemical characterization of a glycosyltransferase Gtf3 from Mycobacterium smegmatis: a case study of improved protein solubilization. 3 Biotech 10, 436 (2020). https://doi.org/10.1007/s13205-020-02431-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02431-x