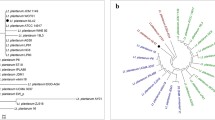
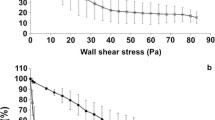
Abstract
Two putative adhesion genes of the potentially probiotic strain Lactobacillus plantarum BFE 5092, i.e., a gene with similarity to an aggregation-promoting factor gene apf5092, and the mucin-binding protein gene mub5092, were investigated in this study. The gene encoding apf5092 encoded a protein bearing a predicted 26 amino acid signal peptide and a LysM domain putatively involved in binding to peptidoglycan. Moreover, the deduced protein also showed an amino acid sequence characteristic of an aggregation-promoting factor. The genes were tested for expression under different environmental conditions, and transcriptional studies on apf5092 showed that the expression could be influenced by low temperature and pH within 30 min. The aggregation behavior of the cells also changed at the low pH condition, but less noticeably at low temperature. To further investigate the role of apf5092 in aggregation, it was cloned and expressed in E. coli. The transformed strain showed higher co-aggregation ability with Gram-positive bacteria. Transcription studies on mub5092 revealed that it could be induced by mucin when added to the growth medium within 30 min. The data suggested that L. plantarum BFE 5092 can quickly adapt to changing environmental conditions and that enhanced aggregation may be important to survive low pH conditions, e.g., of the stomach or of fermented foods, and may thus be an important colonization factor.






Similar content being viewed by others
References
Adlerberth I, Ahrne S, Johansson ML, Molin G, Hanson LA, Wold AE (1996) A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl Environ Microbiol 62:2244–2251
Ahrné S, Lönnermark E, Wold AE, Aberg N, Hesselmar B, Saalman R, Strannegård IL, Molin G, Adlerberth I (2005) Lactobacilli in the intestinal microbiota of Swedish infants. Microbes Infect 7:1256–1262
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Bergonzelli GE, Granato D, Pridmore RD, Marvin-Guy LF, Donnicola D, Corthesy Theulaz IE (2006) GroEL of Lactobacillus johnsonii La1(NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect Immun 74:425–434
Björkroth J, Korkeala H (1996) Evaluation of Lactobacillus sake contamination in vacuum-packaged sliced cooked meat products by ribotyping. J Food Prot 59:398–401
Boekhorst J, Helmer Q, Kleerebezem M, Siezen RJ (2006) Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152:273–280
Bringel F, Castioni A, Olukoya DK, Felis GE, Torriani S, Dellaglio F (2005) Lactobacillus plantarum subsp. argentoratensis subsp. nov., isolated from vegetable matrices. Int J Syst Evol Microbiol 55:1629–1634
Buist G, Steen A, Kok J, Kuipers OP (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847
Bujnakova D, Kmet V (2002) Aggregation of animal lactobacilli with O157 enterohemorrhagic Escherichia coli. J Vet Med B 49:152–154
Castaldo C, Vastano V, Siciliano RA, Candela M, Vici M, Muscariello L, Marasco R, Sacco M (2009) Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb Cell Fact 8:14
Cho GS, Huch M, Hanak A, Holzapfel WH, Franz CMAP (2010) Genetic analysis of the plantaricin EFI locus of Lactobacillus plantarum PCS20 reveals an unusual plantaricin E gene sequence as a result of mutation. Int J Food Microbiol 141:S117–S124
Del Re B, Sgorbati B, Miglioli M, Palenzona D (2000) Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol 31:438–442
Desroche N, Beltramo C, Guzzo J (2005) Determination of an internal control to apply reverse transcription quantitative PCR to study stress response in the lactic acid bacterium Oenococcus oeni. J Microbiol Methods 60:325–333
Desvaux M, Dumas E, Chafsey I, Hébraud M (2006) Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol Lett 256:1–15
de Vries M, Vaughan EE, Kleerebezem M, de Vos WM (2005) Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract. Int Dairy J 16:1018–1028
Goh YJ, Klaenhammer TR (2010) Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl Environ Microbiol 76:5005–5012
Gross G, van der Meulen J, Snel J, van der Meer R, Kleerebezem M, Niewold TA, Hulst MM, Smits MA (2008) Mannose-specific interaction of Lactobacillus plantarum with porcine jejunal epithelium. FEMS Immun Med Microbiol 54:215–223
Gross G, Snel J, Boekhorst J, Smits MA, Kleerebezem M (2010) Biodiversity of mannose-specific adhesion in Lactobacillus plantarum revisited: strain-specific domain composition of the mannose-adhesin. Beneficial Microbes 1:61–66
Gupta V, Garg G (2009) Probiotics. Indian J Med Microbiol 27:202–209
Jankovic I, Venture M, Meylan V, Rouvet M, Elli M, Zink R (2003) Contribution of aggregation-promoting factor (APF) to maintenance of cell shape in Lactobacillus gasseri 4B2. J Bacteriol 185:3288–3296
Jonsson H, Strom E, Roos S (2001) Addition of mucin to the growth medium triggers mucus-binding activity in different strains of Lactobacillus reuteri in vitro. FEMS Microbiol Lett 16:19–22
Kinoshita H, Wakahara N, Watanabe M, Kawasaki T, Matsuo H, Kawai Y, Kitazawa H, Ohnuma S, Miura K, Horii A, Saito T (2008) Cell surface glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of Lactobacillus plantarum LA 318 recognizes human A and B blood group antigens. Res Microbiol 159:685–691
Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ (2003) Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA 100:1990–1995
Kostinek M, Specht I, Edward VA, Schillinger U, Hertel C, Holzapfel WH, Franz CMAP (2005) Diversity and technological properties of predominant lactic acid bacteria from fermented cassava used for preparation of Gari, a traditional African fermented food. Syst Appl Microbiol 28:527–540
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time PCR and the 2(-Delta DeltaC(T)) Method. Methods 25:402–408
Lorca G, Torino MI, Font d V, Ljungh AA (2002) Lactobacilli express cell surface proteins which mediate binding of immobilized collagen and fibronectin. FEMS Microbiol Lett 206:31–37
Marcotte H, Ferrari S, Cesena L, Hammarström L, Morelli L, Pozzi G, Oggioni MR (2004) The aggregation-promoting factor of Lactobacillus crispatus M247 and its genetic locus. J Appl Microbiol 97:749–756
Mathara JM, Schillinger U, Kutima PM, Mbugua SK, Holzapfel WH (2004) Isolation, identification and characterization of the dominant microorganisms of Kule naoto: the Maasai traditional fermented milk in Kenya. Int J Food Microbiol 94:269–278
Mathara JM, Schillinger U, Guigas C, Franz C, Kutima PM, Mbugua SK, Shin HK, Holzapfel WH (2008) Functional characteristics of Lactobacillus ssp. from traditional Maasai fermented milk products in Kenya. Int J Food Microbiol 126:57–64
Mobili P, s Serradell M, Traji SA, Avilés Puigvert FX, Abraham AG, De Antoni GL (2009) Heterogeneity of S-layer proteins from aggregating and non-aggregating Lactobacillus kefir strains. Antonie van Leeuwenhoek 95:363–372
Molenaar D, Bringel F, Schuren FH, de Vos WM, Siezen RJ, Kleerebezem M (2005) Exploring Lactobacillus plantarum genome diversity by using microarrays. J Bacteriol 187:6119–6127
Pitcher DG, Saunders NA, Owen RJ (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156
Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA, Lambert J, de Vos WM, van der Meer R, Smits MA, Kleerebezem M (2005) Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J Bacteriol 187:6128–6136
Proft T, Baker EN (2008) Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Lifesci 66:612–635
Ramiah K, Van Reene A, Dicks LMT (2007) Expression of the mucus adhesion genes mub and mapA, adhesion-like factor EF-Tu and bacteriocin gene plaA of Lactobacillus plantarum 423, monitored with real-time PCR. Int J Food Microbiol 116:405–409
Romero DA, Klaenhammer TR (1992) IS946-mediated integration of heterologous DNA into the genome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol 58:699–702
Sánchez B, Schmitter JM, Urdaci MC (2009) Identification of novel proteins secreted by Lactobacillus plantarum that bind to mucin and fibronectin. J Mol Microbiol Biotechnol 17:158–162
Schachtsiek M, Hammes WP, Hertel C (2004) Characterization of Lactobacillus corynifomis DSM 20001T surface protein Cpf mediating coaggregation with and aggregation among pathogens. Appl Environ Microbiol 70:7078–7085
Siciliano RA, Cacace G, Mazzeo MF, Morelli L, Elli M, Rossi M, Malorni A (2008) Proteomic investigation of the aggregation phenomenon in Lactobacillus crispatus. Biochim Biophys Acta 1784:335–342
Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HT, Rademaker JL, Starrenburg MJ, Kleerebezem M, Molenaar D, van Hylckama Vlieg JE (2010) Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol 12:758–773
Vélez MP, De Keersmaecker SCJ, Vanderleyden J (2007) Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol Lett 276:140–148
Vizoso Pinto GM, Franz CMAP, Schillinger U, Holzapfel WH (2006) Lactobacillus ssp. with in vitro probiotic properties from human faeces and traditional fermented products. Int J Food Microbiol 109:205–214
Vizoso Pinto GM, Schuster T, Riviba K, Watzl B, Holzapfel WH, Franz CMAP (2007) Adhesive and stimulatory properties of potentially probiotic Lactobacillus strains. J Food Prot 70:125–134
Voltan S, Castagliuolo I, Elli M, Longo S, Brun P, D’Incà R, Porzionato GC, Morelli L, Martines D (2007) Aggregating phenotype in Lactobacillus crispatus determines intestinal colonization and TLR2 and TLR4 modulation in murine colonic mucosa. Clin Vaccine Immunol 14:1138–1148
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Grimm, A., Cho, GS., Hanak, A. et al. Characterization of Putative Adhesion Genes in the Potentially Probiotic Strain Lactobacillus plantarum BFE 5092. Probiotics & Antimicro. Prot. 3, 204–213 (2011). https://doi.org/10.1007/s12602-011-9082-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-011-9082-7