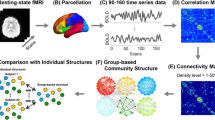
Abstract
The functional system of the human brain can be viewed as a complex network. Among various features of the brain functional network, community structure has raised significant interest in recent years. Increasing evidence has revealed that most realistic complex networks have an overlapping community structure. However, the overlapping community structure of the brain functional network has not been adequately studied. In this paper, we propose a novel method called sparse symmetric non-negative matrix factorization (ssNMF) to detect the overlapping community structure of the brain functional network. Specifically, it is formulated by combining the effective techniques of non-negative matrix factorization and sparse coding. Besides, the non-negative adaptive sparse representation is applied to construct the whole-brain functional network, based on which ssNMF is performed to detect the community structure. Both simulated and real functional magnetic resonance imaging data are used to evaluate ssNMF. The experimental results demonstrate that the proposed ssNMF method is capable of accurately and stably detecting the underlying overlapping community structure. Moreover, the physiological interpretation of the overlapping community structure detected by ssNMF is straightforward. This novel framework, we think, provides an effective tool to study overlapping community structure and facilitates the understanding of the network organization of the functional human brain.










Similar content being viewed by others
References
Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 2009;10(3):186–198.
Girvan M, Newman M. Community structure in social and biological networks. Proc Natl Acad Sci 2002; 99(12):7821–7826.
Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci 2014;17(5): 652–660.
Meunier D, Lambiotte R, Bullmore E. Modular and hierarchically modular organization of brain networks. Front Neurosci 2010;4:200.
Sporns O, Betzel R. Modular brain networks. Annu Rev Psychol 2016;67:613–640.
Ziemke T, Lowe R. On the role of emotion in embodied cognitive architectures: From organisms to robots. Cogn Comput 2009;1(1):104–117.
Yan X. Dissociated emergent response system and fine-processing system in human neural network and a heuristic neural architecture for autonomous humanoid robots. Cogn Comput 2011;3(2):367–373.
Mohan V, Morasso P, Sandini G, Kasderidis S. Inference through embodied simulation in cognitive robots. Cogn Comput 2013;5(3):355–382.
Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. Functional network organization of the human brain. Neuron 2011;72(4):665–678.
Alexander-Bloch A, Lambiotte R, Roberts B, Giedd J, Gogtay N, Bullmore E. The discovery of population differences in network community structure: new methods and applications to brain functional networks in schizophrenia. Neuroimage 2012;59(4):3889–3900.
Thirion B, Dodel S, Poline JB. Detection of signal synchronizations in resting-state fMRI datasets. Neuroimage 2006;29(1):321–327.
van Den Heuvel M, Mandl R, Pol HH. Normalized cut group clustering of resting-state fMRI data. PloS ONE 2008;3(4):e2001.
Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci 2006;103(23):8577–8582.
Von Luxburg U. A tutorial on spectral clustering. Stat Comput 2007;17(4):395–416.
Fortunato S, Hric D. Community detection in networks: a user guide. Phys Rep 2016;659:1–44.
Palla G, Derényi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature 2005;435(7043):814.
Tao D, Li X, Wu X, Maybank S. Geometric mean for subspace selection. IEEE Trans Pattern Anal Mach Intell 2009;31(2):260–274.
Xu C, Tao D, Xu C. Multi-view learning with incomplete views. IEEE Trans Image Process 2015;24 (12):5812–5825.
Liu W, Yang X, Tao D, Cheng J, Tang Y. Multiview dimension reduction via Hessian multiset canonical correlations. Inf Fusion 2018;41:119–128.
Ding C, He X, Simon HD. On the equivalence of non-negative matrix factorization and spectral clustering. In: Proceedings of the 2005 SIAM International Conference on Data Mining; 2005. p. 606–610.
Xie J, Kelley S, Szymanski BK. Overlapping community detection in networks: the state-of-the-art and comparative study. ACM Comput Surv 2013;45(4):43.
Zarei M, Izadi D, Samani KA. Detecting overlapping community structure of networks based on vertex–vertex correlations. J Stat Mech-Theory Exp 2009;11:P11013.
Psorakis I, Roberts S, Ebden M, Sheldon B. Overlapping community detection using bayesian non-negative matrix factorization. Phys Rev E 2011;83(6):066114.
Yang J, Leskovec J. Overlapping community detection at scale: a non-negative matrix factorization approach. In: Proceedings of the sixth ACM International Conference on Web Search and Data Mining; 2013. p. 587–596.
Wang F, Li T, Wang X, Zhu S, Ding C. Community discovery using non-negative matrix factorization. Data Min Knowl Discov 2011;22(3):493–521.
Li X, Hu Z, Wang H. Overlapping community structure detection of brain functional network using non-negative matrix factorization. In: International Conference on Neural Information Processing (ICONIP); 2016. p. 140–147.
Chen S, Xin Y, Luo B. Action-based pedestrian identification via hierarchical matching pursuit and order preserving sparse coding. Cogn Comput 2016;8(5):797–805.
Lv L, Zhao D, Deng Q. A semi-supervised predictive sparse decomposition based on task-driven dictionary learning. Cogn Comput 2017;9(1):115–124.
Liu M, Xu C, Luo Y, Xu C, Wen Y, Tao D. Cost-sensitive feature selection by optimizing F-measures. IEEE Trans Image Process 2018;27(3):1323–1335.
Li Y, Yu Z, Bi N, Xu Y, Gu Z, Amari S. Sparse representation for brain signal processing: a tutorial on methods and applications. IEEE Signal Process Mag 2014;31(3):96–106.
Xie J, Douglas PK, Wu YN, Brody AL, Anderson AE. Decoding the encoding of functional brain networks: an fMRI classification comparison of non-negative matrix factorization (NMF), independent component analysis (ICA), and sparse coding algorithms. J Neurosci Method 2017;282:81–94.
Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 1995;34(4):537–541.
Li X, Hu Z, Wang H. Sparse-network based framework for detecting the overlapping community structure of brain functional network. In: Advances in Brain Inspired Cognitive Systems; 2016. p. 355–365.
Lee D, Seung H. Learning the parts of objects by non-negative matrix factorization. Nature 1999;401(6755): 788–791.
Guan N, Tao D, Luo Z, Shawe-Taylor J. 2012. MahNMF: Manhattan non-negative matrix factorization. arXiv:1207.3438.
Liu T, Gong M, Tao D. Large-cone non-negative matrix factorization. IEEE Trans Neural Netw Learn Syst 2017;28(9):2129– 2142.
Shan D, Xu X, Liang T, Ding S. Rank-adaptive non-negative matrix factorization. Cogn Comput. 2018;10(3):506–515.
Padilla P, López M, Górriz J M, Ramirez J, Salas-Gonzalez D, Álvarez I. NMF-SVM based CAD tool applied to functional brain images for the diagnosis of Alzheimer’s disease. IEEE Trans Med Imaging 2012; 31(2):207–216.
Wang Y, Zhang Y. Non-negative matrix factorization: A comprehensive review. IEEE Trans Knowl Data Eng 2013;25(6):1336–1353.
Zhang Z, Wang Y, Ahn YY. Overlapping community detection in complex networks using symmetric binary matrix factorization. Phys Rev E 2013;87(6):062803.
Zhang Y, Yeung DY. Overlapping community detection via bounded non-negative matrix tri-factorization. In: Proceedings of the 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2012. p. 606–614.
Zhang Z, Xu Y, Yang J, Li X, Zhang D. A survey of sparse representation: algorithms and applications. IEEE Access 2015;3:490–530.
Donoho DL, Elad M. Optimally sparse representation in general (non-orthogonal) dictionaries via ℓ 1 minimization. Proc Natl Acad Sci 2003;100(5):2197–2202.
Guo S, Wang Z, Ruan Q. Enhancing sparsity via ℓ p(0 < p < 1) minimization for robust face recognition. Neurocomputing 2013;99:592–602.
Bengio S, Pereira F, Singer Y, Strelow D. Group sparse coding. In: Advances in Neural Information Processing Systems; 2009. p. 82–89.
Liu W, Zha Z, Wang Y, Lu K, Tao D. P-Laplacian regularized sparse coding for human activity recognition. IEEE Trans Ind Electron 2016;63(8):5120–5129.
Hoyer PO. Non-negative sparse coding. In: Proceedings of the 12th IEEE Workshop on Neural Networks for Signal Processing; 2002. p. 557–565.
Hoyer PO. Non-negative matrix factorization with sparseness constraints. J Mach Learn Res 2004;5(Nov): 1457–1469.
Gao Y, Church G. Improving molecular cancer class discovery through sparse non-negative matrix factorization. Bioinformatics 2005;21(21):3970–3975.
Grave E, Obozinski GR. Bach FR. Trace lasso: a trace norm regularization for correlated designs. In: Advances in Neural Information Processing Systems; 2011. p. 2187–2195.
Lu C, Feng J, Lin Z, Yan S. Correlation adaptive subspace segmentation by trace lasso. In: IEEE International Conference on Computer Vision; 2013. p. 1345–1352.
Li X, Wang H. Identification of functional networks in resting state fMRI data using adaptive sparse representation and affinity propagation clustering. Front Neurosci 2015;9:383.
Lin CJ. On the convergence of multiplicative update algorithms for non-negative matrix factorization. IEEE Trans Neural Netw 2007;18(6):1589–1596.
Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci 2008;105(4):1118–1123.
Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech 2008;2008(10):P10008.
Shi J, Malik J. Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 2000;22(8): 888–905.
Frey BJ, Dueck D. Clustering by passing messages between data points. Science 2007;315(5814):972–976.
Eavani H, Satterthwaite TD, Filipovych R, Gur RE, Gur RC, Davatzikos C. Identifying sparse connectivity patterns in the brain using resting-state fMRI. Neuroimage 2015;105:286– 299.
Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for fMRI. Neuroimage 2011;54(2):875–891.
Lovász L, Plummer MD. Matching theory. Ann Discret Math. 1986;29:1–543.
Yang J, Leskovec J. Overlapping community detection at scale: a nonnegative matrix factorization approach. In: Proceedings of the 6th ACM International Conference on Web Search and Data Mining; 2013. p. 587–596.
McDaid AF, Greene D, Hurley N. 2011. Normalized mutual information to evaluate overlapping community finding algorithms. arXiv:1110.2515.
Gregory S. Fuzzy overlapping communities in networks. J Stat Mech: Theory Exp 2011;2011(02):P02017.
Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, et al. Toward discovery science of human brain function. Proc Natl Acad Sci 2010;107(10):4734–4739.
Zuo XN, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J, et al. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data 2014;1:140049.
Chen B, Xu T, Zhou C, Wang L, Yang N, Wang Z. Individual variability and test–retest reliability revealed by ten repeated resting-state brain scans over one month. PloS ONE 2015;10:e0144963.
Yan C, Zang Y. DPARSF: A matlab toolbox for “pipeline” data analysis of resting-state fMRI. Front Syst Neurosci 2010;4:13.
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15(1):273–289.
Campello RJGB, Hruschka ER. A fuzzy extension of the silhouette width criterion for cluster analysis. Fuzzy Set Syst 2006;157(21):2858–2875.
Fransson P, Marrelec G. The precuneus/posterior cingulate cortex plays a pivotal role in the default mode network: evidence from a partial correlation network analysis. Neuroimage 2008;42:1178–1184.
van den Heuvel MP, Pol HEH. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol 2010;20(8):519–534.
Isaacson R. The limbic system. Berlin: Springer Science & Business Media; 2013.
Cole MW, Reynolds JR, Power JD, Repovs G, Anticevic A, Braver TS. Multi-task connectivity reveals flexible hubs for adaptive task control. Nature Neurosci 2013;16(9):1348– 1355.
Mazoyer B, Zago L, Mellet E, Bricogne S, Etard O, Houdé O, et al. Cortical networks for working memory and executive functions sustain the conscious resting state in man. Brain Res Bull 2001;54(3):287–298.
Yeo BTT, Krienen FM, Chee MWL, Buckner RL. Estimates of segregation and overlap of functional connectivity networks in the human cerebral cortex. Neuroimage 2014;88:212–227.
van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn Sci 2013;17:683–696.
Peharz R, Pernkopf F. Sparse nonnegative matrix factorization with ℓ 0-constraints. Neurocomputing 2012;80:38–46.
Kong D, Fujimaki R, Liu J, Nie F, Ding C. Exclusive feature learning on arbitrary structures via ℓ 1,2-norm. In: Advances in Neural Information Processing Systems; 2014. p. 1655–1663.
He Z, Xie S, Zdunek R, Zhou G, Cichocki A. Symmetric nonnegative matrix factorization: Algorithms and applications to probabilistic clustering. IEEE Trans Neural Netw 2011;22(12):2117–2131.
Kuang D, Ding C, Park H. Symmetric nonnegative matrix factorization for graph clustering. In: Proceedings of the 2012 SIAM International Conference on Data Mining; 2012. p. 106–117.
Huang K, Sidiropoulos ND, Swami A. Non-negative matrix factorization revisited: Uniqueness and algorithm for symmetric decomposition. IEEE Trans Signal Process 2014; 62 (1): 211– 224.
Acknowledgements
The authors wish to thank the editors and reviewers for the comments and recommendations, which have helped improve the paper substantially.
Funding
This work was supported in part by the National Natural Science Foundation of China under grants 61773114 and 61472089, the Joint Fund of the National Natural Science Foundation of China and Guangdong Province under grant U1501254, the Science and Technology Planning Project of Guangdong Province under grants 2015B010131015 and 2015B010108006, Key Project of Internation as well as Hongkong, Macao & Taiwan Innovation Platform and International Cooperation by Universities in Guangdong Province under grant 2015KGJHZ023, and the China Scholarship Council Fund under Grant 201603780037.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval: All the procedures performed in the studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee as well as with the 1964 Helsinki declaration and its later amendments, or comparable ethical standards.
Conflict of interests
The authors declare that they have no conflict of interest.
Informed Consent
Informed consent was obtained from all the individual participants included in the study.
Rights and permissions
About this article
Cite this article
Li, X., Hu, Z. & Wang, H. Combining Non-negative Matrix Factorization and Sparse Coding for Functional Brain Overlapping Community Detection. Cogn Comput 10, 991–1005 (2018). https://doi.org/10.1007/s12559-018-9585-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12559-018-9585-6