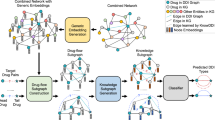
Abstract
The search for potential drug–disease associations (DDA) can speed up drug development cycles, reduce costly wasted resources, and accelerate disease treatment by repurposing existing drugs that can control further disease progression. As technologies such as deep learning continue to mature, many researchers tend to use emerging technologies to predict potential DDA. The performance of DDA prediction is still challenging and there is some space for improvement due to issues such as the small number of existing associations and possible noise in the data. To better predict DDA, we propose a computational approach based on hypergraph learning with subgraph matching (HGDDA). In particular, HGDDA first extracts feature subgraph information in the validated drug–disease association network and proposes a negative sampling strategy based on similarity network to reduce the data imbalance. Second, the hypergraph Unet module is used by extracting Finally, the potential DDA is predicted by designing a hypergraph combination module to convolution and pooling the two constructed hypergraphs separately, and calculating the difference information between the subgraphs using cosine similarity for node matching. The performance of HGDDA is verified under two standard datasets by 10-fold cross-validation (10-CV), and the results outperform existing drug–disease prediction methods. In addition, to validate the overall utility of the model, the top 10 drugs for the specific disease are predicted through the case study and validated using the CTD database.
Graphical abstract






Similar content being viewed by others
References
Chan HS, Shan H, Dahoun T, Vogel H, Yuan S (2019) Advancing drug discovery via artificial intelligence. Trends Pharmacol Sci 40(8):592–604. https://doi.org/10.1016/j.tips.2019.06.004
Baudot A, Gomez-Lopez G, Valencia A (2009) Translational disease interpretation with molecular networks. Genome Biol 10(6):1–9. https://doi.org/10.1186/gb-2009-10-6-221
Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabási A-L (2007) The human disease network. Proc Natl Acad Sci 104(21):8685–8690. https://doi.org/10.1073/pnas.0701361104
Luo H, Wang J, Li M, Luo J, Peng X, Wu F-X, Pan Y (2016) Drug repositioning based on comprehensive similarity measures and bi-random walk algorithm. Bioinformatics 32(17):2664–2671. https://doi.org/10.1093/bioinformatics/btw228
Wang W, Yang S, Zhang X, Li J (2014) Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics 30(20):2923–2930. https://doi.org/10.1093/bioinformatics/btu403
Wang Y, Guo M, Ren Y, Jia L, Yu G (2019) Drug repositioning based on individual bi-random walks on a heterogeneous network. BMC Bioinform 20(15):1–13. https://doi.org/10.1186/s12859-019-3117-6
Xuan P, Cui H, Shen T, Sheng N, Zhang T (2019) Heterodualnet: a dual convolutional neural network with heterogeneous layers for drug–disease association prediction via chou’s five-step rule. Front Pharmacol 10:1301. https://doi.org/10.3389/fphar.2019.01301
Jiang H-J, Huang Y-A, You Z-H (2019) Predicting drug-disease associations via using gaussian interaction profile and kernel-based autoencoder. BioMed Res Int. https://doi.org/10.1155/2019/2426958
Wang Y, Deng G, Zeng N, Song X, Zhuang Y (2019) Drug–disease association prediction based on neighborhood information aggregation in neural networks. IEEE Access 7:50581–50587. https://doi.org/10.1109/ACCESS.2019.2907522
Kitsiranuwat S, Suratanee A, Plaimas K (2021) Multi-data aspects of protein similarity with a learning technique to identify drug–disease associations. Appl Sci 11(7):2914. https://doi.org/10.3390/app11072914
Kitsiranuwat S, Suratanee A, Plaimas K (2022) Integration of various protein similarities using random forest technique to infer augmented drug-protein matrix for enhancing drug–disease association prediction. Sci Prog 105(3):00368504221109215. https://doi.org/10.1177/00368504221109215
Kipf TN, Welling M (2016) Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907. https://doi.org/10.48550/arXiv.1609.02907
Yu Z, Huang F, Zhao X, Xiao W, Zhang W (2021) Predicting drug–disease associations through layer attention graph convolutional network. Brief Bioinform 22(4):243. https://doi.org/10.1093/bib/bbaa243
Cai L, Lu C, Xu J, Meng Y, Wang P, Fu X, Zeng X, Su Y (2021) Drug repositioning based on the heterogeneous information fusion graph convolutional network. Brief Bioinform 22(6):319. https://doi.org/10.1093/bib/bbab319
Gottlieb A, Stein GY, Ruppin E, Sharan R (2011) Predict: a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol 7(1):496. https://doi.org/10.1038/msb.2011.26
Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M (2008) Drugbank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36(suppl 1):901–906. https://doi.org/10.1093/nar/gkm958
Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA (2000) Online mendelian inheritance in man (omim). Hum Mutat 15(1):57–61. https://doi.org/10.1002/ajmg.a.62407
Van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JA (2006) A text-mining analysis of the human phenome. Eur J Hum Genet 14(5):535–542. https://doi.org/10.1038/sj.ejhg.5201585
Hu P, Huang Y-A, Mei J, Leung H, Chen Z-H, Kuang Z-M, You Z-H, Hu L (2021) Learning from low-rank multimodal representations for predicting disease–drug associations. BMC Med Inform Decision Making 21(1):1–13. https://doi.org/10.1186/s12911-021-01648-x
Gleich DF (2015) Pagerank beyond the web. SIAM Rev 57(3):321–363. https://doi.org/10.1137/140976649
Gao H, Ji S (2019) Graph u-nets. In: International conference on machine learning, PMLR, pp 2083–2092. https://doi.org/10.1109/TPAMI.2021.3081010
Feng Y, You H, Zhang Z, Ji R, Gao Y (2019) Hypergraph neural networks. In: Proceedings of the AAAI conference on artificial intelligence, vol 33, pp 3558–3565. https://doi.org/10.1609/aaai.v33i01.33013558
Guo G, Wang H, Bell D, Bi Y, Greer K (2003) Knn model-based approach in classification. In: OTM confederated international conferences on the move to meaningful internet systems. Springer, pp 986–996. https://doi.org/10.1007/978-3-540-39964-3_62
Li R-H, Yu JX, Qin L, Mao R, Jin T (2015) On random walk based graph sampling. In: 2015 IEEE 31st international conference on data engineering. IEEE, pp 927–938. https://doi.org/10.1109/ICDE.2015.7113345
Diehl F (2019) Edge contraction pooling for graph neural networks. arXiv preprint arXiv:1905.10990. https://doi.org/10.48550/arXiv.1905.10990
Feng F, He X, Zhang H, Chua T-S (2021) Cross-gcn: enhancing graph convolutional network with k-order feature interactions. IEEE Trans Knowl Data Eng. https://doi.org/10.1109/TKDE.2021.3077524
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980. https://doi.org/10.48550/arXiv.1412.6980
Mongia A, Chouzenoux E, Majumdar A (2022) Computational prediction of drug–disease association based on graph-regularized one bit matrix completion. IEEE/ACM Trans Comput Biol Bioinf. https://doi.org/10.1109/TCBB.2022.3189879
Luo H, Li M, Wang S, Liu Q, Li Y, Wang J (2018) Computational drug repositioning using low-rank matrix approximation and randomized algorithms. Bioinformatics 34(11):1904–1912. https://doi.org/10.1093/bioinformatics/bty013
Yang M, Luo H, Li Y, Wang J (2019) Drug repositioning based on bounded nuclear norm regularization. Bioinformatics 35(14):455–463. https://doi.org/10.1093/bioinformatics/btz331
Davis AP, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, Wiegers TC, Mattingly CJ (2021) Comparative toxicogenomics database (ctd): update 2021. Nucleic Acids Res 49(D1):1138–1143. https://doi.org/10.1093/nar/gkaa891
Funding
This research is supported by Scientific Research Fund Project of the Education Department of Liaoning Province (No. LJKZ0028).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Song, J., Wei, M. et al. Predicting Potential Drug–Disease Associations Based on Hypergraph Learning with Subgraph Matching. Interdiscip Sci Comput Life Sci 15, 249–261 (2023). https://doi.org/10.1007/s12539-023-00556-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-023-00556-0