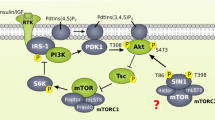
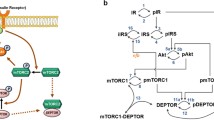
Abstract
Mechanistic target of rapamycin (mTOR) is a critical protein in the regulation of cell fate decision making, especially in cancer cells. mTOR acts as a signal integrator and is one of the main elements of interactions among the pivotal cellular processes such as cell death, autophagy, metabolic reprogramming, cell growth, and cell cycle. The temporal control of these processes is essential for the cellular homeostasis and dysregulation of mTOR signaling pathway results in different phenotypes, including aging, oncogenesis, cell survival, cell growth, senescence, quiescence, and cell death. In this paper, we have proposed a systems biology roadmap to study mTOR control system, which introduces the theoretical and experimental modalities to decode temporal and dynamical characteristics of mTOR signaling in cancer.




Similar content being viewed by others
References
Sabatini DM (2006) mTOR and cancer: insights into a complex relationship. Nat Rev Cancer 6(9):729
Laplante M, Sabatini DM (2009) mTOR signaling at a glance. J Cell Sci 122(20):3589–3594
Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149(2):274–293
Inoki K, Corradetti MN, Guan K-L (2005) Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet 37(1):19
Guertin DA, Sabatini DM (2009) The pharmacology of mTOR inhibition. Sci Signal 2(67):pe24-pe24
Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168(6):960–976
Tyson JJ, Baumann WT, Chen C, Verdugo A, Tavassoly I, Wang Y, Weiner LM, Clarke R (2011) Dynamic modelling of oestrogen signalling and cell fate in breast cancer cells. Nat Rev Cancer 11(7):523
Dann SG, Selvaraj A, Thomas G (2007) mTOR Complex1–S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med 13(6):252–259
Vergès B, Cariou B (2015) mTOR inhibitors and diabetes. Diabetes Res Clin Pract 110(2):101–108
Jia G, Aroor AR, Martinez-Lemus LA, Sowers JR (2014) Overnutrition, mTOR signaling, and cardiovascular diseases. Am J Physiol Regul Integr Comp Physiol 307(10):R1198–R1206
Perl A (2016) Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat Rev Rheumatol 12(3):169
Ginzberg MB, Chang N, D’Souza H, Patel N, Kafri R, Kirschner MW (2018) Cell size sensing in animal cells coordinates anabolic growth rates and cell cycle progression to maintain cell size uniformity. Elife 7:e26957
Xu S, Cai Y, Wei Y (2014) mTOR signaling from cellular senescence to organismal aging. Aging Dis 5(4):263
Lamming DW, Ye L, Sabatini DM, Baur JA (2013) Rapalogs and mTOR inhibitors as anti-aging therapeutics. J Clin Investig 123(3):980–989
Ginzberg MB, Kafri R, Kirschner M (2015) On being the right (cell) size. Science 348(6236):1245075
Garratt M, Nakagawa S, Simons MJ (2016) Comparative idiosyncrasies in life extension by reduced mTOR signalling and its distinctiveness from dietary restriction. Aging Cell 15(4):737–743
Powell JD, Pollizzi KN, Heikamp EB, Horton MR (2012) Regulation of immune responses by mTOR. Annu Rev Immunol 30:39–68
Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R (2009) mTOR regulates memory CD8 T-cell differentiation. Nature 460(7251):108
Mannick JB, Morris M, Hockey HUP, Roma G, Beibel M, Kulmatycki K, Watkins M, Shavlakadze T, Zhou W, Quinn D et al (2018) TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci Transl Med 10(449):ee1564
Huang S, Houghton PJ (2003) Targeting mTOR signaling for cancer therapy. Curr Opin Pharmacol 3(4):371–377
Faivre S, Kroemer G, Raymond E (2006) Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov 5(8):671
Benjamin D, Colombi M, Moroni C, Hall MN (2011) Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov 10(11):868
Neuhaus P, Klupp J, Langrehr JM (2001) mTOR inhibitors: an overview. Liver Transpl 7(6):473–484
Vezina C, Kudelski A, Sehgal S (1975) Rapamycin (AY-22, 989), a new antifungal antibiotic. J Antibiot 28(10):721–726
Janku F, Yap TA, Meric-Bernstam F (2018) Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 15(5):273
Porta C, Paglino C, Mosca A (2014) Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol 4:64
Sahra IB, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, Tanti J-F, Giorgetti-Peraldi S, Bost F (2011) Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Can Res 71(13):4366–4372
Tavassoly I, Goldfarb J, Iyengar R (2018) Systems biology primer: the basic methods and approaches. Essays Biochem 62(4):487–500
Din FVN, Valanciute A, Houde VP, Zibrova D, Green KA, Sakamoto K, Alessi DR, Dunlop MG (2012) Aspirin inhibits mTOR signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology 142(7):1504–1515.e1503
Shimobayashi M, Hall MN (2014) Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 15(3):155
Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128(4):683–692
Hansen J, Meretzky D, Woldesenbet S, Stolovitzky G, Iyengar R (2017) A flexible ontology for inference of emergent whole cell function from relationships between subcellular processes. Sci Rep 7(1):17689
Clarke R, Shajahan AN, Wang Y, Tyson JJ, Riggins RB, Weiner LM, Bauman WT, Xuan J, Zhang B, Facey C (2011) Endoplasmic reticulum stress, the unfolded protein response, and gene network modeling in antiestrogen resistant breast cancer. Horm Mol Biol Clin Investig 5(1):35–44
Clarke R, Cook KL, Hu R, Facey CO, Tavassoly I, Schwartz JL, Baumann WT, Tyson JJ, Xuan J, Wang Y (2012) Endoplasmic reticulum stress, the unfolded protein response, autophagy, and the integrated regulation of breast cancer cell fate. Can Res 72(6):1321–1331
Barabási A-L, Gulbahce N, Loscalzo J (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12(1):56
Dorvash M, Farahmandnia M, Mosaddeghi P, Farahmandnejad M, Saber H, Khorraminejad-Shirazi M, Azadi A, Tavassoly I (2019) Dynamic modeling of signal transduction by mTOR complexes in cancer. J Theor Biol 483:109992
Meng D, Frank AR, Jewell JL (2018) mTOR signaling in stem and progenitor cells. Development 145(1):dev152595
Tavassoly I, Parmar J, Shajahan-Haq A, Clarke R, Baumann W, Tyson J (2015) Dynamic modeling of the interaction between autophagy and apoptosis in mammalian cells. CPT Pharmacomet Syst Pharmacol 4(4):263–272
Tavassoly I (2015) Dynamics of cell fate decision mediated by the interplay of autophagy and apoptosis in cancer cells mathematical modeling and experimental observations. Springer, New York
Parmar JH, Cook KL, Shajahan-Haq AN, Clarke PA, Tavassoly I, Clarke R, Tyson JJ, Baumann WT (2013) Modelling the effect of GRP78 on anti-oestrogen sensitivity and resistance in breast cancer. Interface Focus 3(4):20130012
Shaw RJ, Cantley LC (2006) Ras, PI (3) K and mTOR signalling controls tumour cell growth. Nature 441(7092):424
Chatterjee A (2015) Control of cell cycle progression by mTOR
Kim J, Guan K-L (2019) mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol 21(1):63
Efeyan A, Sabatini DM (2010) mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol 22(2):169–176
Rosner M, Fuchs C, Siegel N, Valli A, Hengstschläger M (2009) Functional interaction of mammalian target of rapamycin complexes in regulating mammalian cell size and cell cycle. Hum Mol Genet 18(17):3298–3310
Nishitani S, Horie M, Ishizaki S, Yano H (2013) Branched chain amino acid suppresses hepatocellular cancer stem cells through the activation of mammalian target of rapamycin. PLoS One 8(11):e82346
Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R (2012) TGF-β-induced activation of mTOR complex 2 drives epithelial–mesenchymal transition and cell invasion. J Cell Sci 125(5):1259–1273
Loos B, Engelbrecht A-M (2009) Cell death: a dynamic response concept. Autophagy 5(5):590–603
Laplante M, Sabatini DM (2012) mTOR signaling. Cold Spring Harb Perspect Biol 4(2):a011593
Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485(7396):109
Domhan S, Schwager C, Wei Q, Muschal S, Sommerer C, Morath C, Wick W, Maercker C, Debus J, Zeier M (2014) Deciphering the systems biology of mTOR inhibition by integrative transcriptome analysis. Curr Pharm Des 20(1):88–100
Fu Y, Zheng X, Jia X, Binderiya U, Wang Y, Bao W, Bao L, Zhao K, Fu Y, Hao H (2016) A quantitative transcriptomic analysis of the physiological significance of mTOR signaling in goat fetal fibroblasts. BMC Genomics 17(1):879
Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485(7396):55
Hsieh H-J, Zhang W, Lin S-H, Yang W-H, Wang J-Z, Shen J, Zhang Y, Lu Y, Wang H, Yu J (2018) Systems biology approach reveals a link between mTORC1 and G2/M DNA damage checkpoint recovery. Nat Commun 9(1):3982
Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, Perreault C, Roux PP, Kitano H (2010) A comprehensive map of the mTOR signaling network. Mol Syst Biol 6(1):453
Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma’ayan A (2011) Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics 28(1):105–111
Sorribes I, Basu A, Brady R, Enriquez-Navas P, Feng X, Kather J, Nerlakanti N, Stephens R, Strobl M, Tavassoly I (2019) Harnessing patient-specific response dynamics to optimize evolutionary therapies for metastatic clear cell renal cell carcinoma-learning to adapt. bioRxiv, 563130. https://doi.org/10.1101/563130
Rockne RC, Hawkins-Daarud A, Swanson KR, Sluka JP, Glazier JA, Macklin P, Hormuth DA, Jarrett AM, Lima EABF, Tinsley Oden J et al (2019) The 2019 mathematical oncology roadmap. Phys Biol 16(4):041005
Bibi Z, Ahmad J, Siddiqa A, Paracha RZ, Saeed T, Ali A, Janjua HA, Ullah S, Ben Abdallah E, Roux O (2017) Formal modeling of mTOR associated biological regulatory network reveals novel therapeutic strategy for the treatment of cancer. Front Physiol 8:416
Sulaimanov N, Klose M, Busch H, Boerries M (2017) Understanding the mTOR signaling pathway via mathematical modeling. Wiley Interdiscip Rev Syst Biol Med 9(4):e1379
Tyson JJ, Chen K, Novak B (2001) Network dynamics and cell physiology. Nat Rev Mol Cell Biol 2(12):908
Wang R-S, Saadatpour A, Albert R (2012) Boolean modeling in systems biology: an overview of methodology and applications. Phys Biol 9(5):055001
Albert I, Thakar J, Li S, Zhang R, Albert R (2008) Boolean network simulations for life scientists. Source Code Biol Med 3(1):16
Zadeh LA (1965) Fuzzy sets. Inf Control 8(3):338–353
Dalle Pezze P, Sonntag AG, Thien A, Prentzell MT, Gödel M, Fischer S, Neumann-Haefelin E, Huber TB, Baumeister R, Shanley DP (2012) A dynamic network model of mTOR signaling reveals TSC-independent mTORC2 regulation. Sci Signal 5(217):ra25–ra25
Sonntag AG, Dalle Pezze P, Shanley DP, Thedieck K (2012) A modelling–experimental approach reveals insulin receptor substrate (IRS)-dependent regulation of adenosine monosphosphate-dependent kinase (AMPK) by insulin. FEBS J 279(18):3314–3328
Kriete A, Bosl WJ, Booker G (2010) Rule-based cell systems model of aging using feedback loop motifs mediated by stress responses. PLoS Comput Biol 6(6):e1000820
Wu M, Yang X, Chan C (2009) A dynamic analysis of IRS-PKR signaling in liver cells: a discrete modeling approach. PLoS One 4(12):e8040
Dalle Pezze P, Ruf S, Sonntag AG, Langelaar-Makkinje M, Hall P, Heberle AM, Navas PR, Van Eunen K, Tölle RC, Schwarz JJ (2016) A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat Commun 7:13254
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K (2016) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 12(1):1–222
Maiuri MC, Zalckvar E, Kimchi A, Kroemer G (2007) Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 8(9):741
Jung CH, Seo M, Otto NM, Kim D-H (2011) ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy 7(10):1212–1221
Blagosklonny MV (2008) Aging, stem cells, and mammalian target of rapamycin: a prospect of pharmacologic rejuvenation of aging stem cells. Rejuvenation Res 11(4):801–808
Novák B, Tyson JJ (2008) Design principles of biochemical oscillators. Nat Rev Mol Cell Biol 9(12):981
Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: simple building blocks of complex networks. Science 298(5594):824–827
Tyson JJ, Novák B (2010) Functional motifs in biochemical reaction networks. Annu Rev Phys Chem 61:219–240
Tian X-J, Zhang X-P, Liu F, Wang W (2009) Interlinking positive and negative feedback loops creates a tunable motif in gene regulatory networks. Phys Rev E 80(1):011926
Tsai TY-C, Choi YS, Ma W, Pomerening JR, Tang C, Ferrell JE (2008) Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science 321(5885):126–129
Nazio F, Cecconi F (2017) Autophagy up and down by outsmarting the incredible ULK. Autophagy 13(5):967–968
Lamming DW, Bar-Peled L (2019) Lysosome: the metabolic signaling hub. Traffic 20(1):27–38
Ryzhikov M, Ehlers A, Steinberg D, Xie W, Oberlander E, Brown S, Gilmore PE, Townsend RR, Lane WS, Dolinay T (2019) Diurnal rhythms spatially and temporally organize autophagy. Cell Rep 26(7):1880–1892.e1886
Szymańska P, Martin KR, MacKeigan JP, Hlavacek WS, Lipniacki T (2015) Computational analysis of an autophagy/translation switch based on mutual inhibition of MTORC1 and ULK1. PLoS One 10(3):e0116550
Sachdeva UM, Thompson CB (2008) Diurnal rhythms of autophagy: implications for cell biology and human disease. Autophagy 4(5):581–589
Ma D, Panda S, Lin JD (2011) Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J 30(22):4642–4651
Nazio F, Carinci M, Valacca C, Bielli P, Strappazzon F, Antonioli M, Ciccosanti F, Rodolfo C, Campello S, Fimia GM (2016) Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J Cell Biol 215(6):841–856
Cao R, Obrietan K (2010) mTOR signaling and entrainment of the mammalian circadian clock. Mol Cell Pharmacol 2(4):125
Ramanathan C, Kathale ND, Liu D, Lee C, Freeman DA, Hogenesch JB, Cao R, Liu AC (2018) mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet 14(5):e1007369
Toledo M, Batista-Gonzalez A, Merheb E, Aoun ML, Tarabra E, Feng D, Sarparanta J, Merlo P, Botrè F, Schwartz GJ (2018) Autophagy regulates the liver clock and glucose metabolism by degrading CRY1. Cell Metab 28(2):268–281.e264
Ma D, Li S, Molusky MM, Lin JD (2012) Circadian autophagy rhythm: a link between clock and metabolism? Trends Endocrinol Metab 23(7):319–325
Devalla HD, Schwach V, Ford JW, Milnes JT, El-Haou S, Jackson C, Gkatzis K, Elliott DA, de Sousa Lopes SMC, Mummery CL (2015) Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med 7(4):394–410
Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 13(4):453
Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ (2009) Autophagy regulates lipid metabolism. Nature 458(7242):1131
Maiese K (2017) Moving to the rhythm with clock (circadian) genes, autophagy, mTOR, and SIRT1 in degenerative disease and cancer. Curr Neurovasc Res 14(3):299–304
Savvidis C, Koutsilieris M (2012) Circadian rhythm disruption in cancer biology. Mol Med 18(9):1249–1260
Wang S, Tsun Z-Y, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W (2015) Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347(6218):188–194
López-Otín C, Galluzzi L, Freije JM, Madeo F, Kroemer G (2016) Metabolic control of longevity. Cell 166(4):802–821
Rubinsztein DC, Mariño G, Kroemer G (2011) Autophagy and aging. Cell 146(5):682–695
Leontieva OV, Demidenko ZN, Gudkov AV, Blagosklonny MV (2011) Elimination of proliferating cells unmasks the shift from senescence to quiescence caused by rapamycin. PLoS One 6(10):e26126
Mathiassen SG, De Zio D, Cecconi F (2017) Autophagy and the cell cycle: a complex landscape. Front Oncol 7:51
Kriel J, Loos B (2019) The good, the bad and the autophagosome: exploring unanswered questions of autophagy-dependent cell death. Cell Death Differ 26(4):640–652
Tavassoly I, Hu Y, Zhao S, Mariottini C, Boran A, Chen Y, Li L, Tolentino RE, Jayaraman G, Goldfarb J et al (2019) Genomic signatures defining responsiveness to allopurinol and combination therapy for lung cancer identified by systems therapeutics analyses. Mol Oncol 13(8):1725–1743
Zaytseva YY, Valentino JD, Gulhati P, Evers BM (2012) mTOR inhibitors in cancer therapy. Cancer Lett 319(1):1–7
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dorvash, M., Farahmandnia, M. & Tavassoly, I. A Systems Biology Roadmap to Decode mTOR Control System in Cancer. Interdiscip Sci Comput Life Sci 12, 1–11 (2020). https://doi.org/10.1007/s12539-019-00347-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-019-00347-6