Abstract
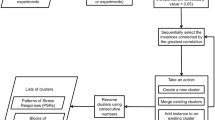
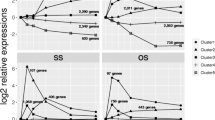
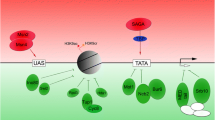
For living organisms like Saccharomyces cerevisiae, instinctual response to sudden environmental changes leads to swift establishment of adaptive mechanisms. The issue about how TFs sense and adapt to various environmental stresses has not been systematically studied yet. Here we try to elucidate this problem with the assistance of genomic expression patterns from a computation perspective. A dynamic transcriptional regulatory model is employed to uncover significant TF-target regulatory relationships under various environmental stresses. Based on a global microarray dataset that describes how transcriptional regulators significantly respond to one specific stress, we constructed a sensory transcriptional network for the potential specific stressresponsive regulators. Alternatively, we have observed cross-talks among these sensory transcription networks that may shed light on general stress-responsive regulators. Results reveal that our method not only reconstructs the potential global protection mechanisms under various environmental stresses but also presents a set of reported specific stress-responsive regulators (i.e., Aft2, Hsf1, Msn2, Msn4, Skn7 and Yap1) as well as a set of inferred specific/general stress-responsive regulators that may provide new guidance for further experiments on yeast cells’ adaption to environmental stimuli. Though we only make a study on the yeast S. cerevisiae, our method can be broadly applied to all species.
Similar content being viewed by others
References
Alon, U. 2007. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall/CRC, New York.
Blaiseau, P.L., Lesuisse, E., Camadro, J.M. 2001. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J Biol Chem 276, 34221–6.
Causton, H.C., Ren, B., Koh, S.S. et al. 2001. Remodeling of yeast genome expression in response to environmental changes. Molecular Biology of the Cell 12, 323–337.
Chen, H.C., Lee, H.C., Lin, T.Y. et al. 2004. Quantitative characterization of the transcriptional regulatory network in the yeast cell cycle. Bioinformatics 20, 1914–1927.
Chen, K.C., Wang, T.Y., Tseng, H.H. et al. 2005. A stochastic differential equation model for quantifying transcriptional regulatory network in Saccharomyces cerevisiae. Bioinformatics 21, 2883–2890.
De Nadal, E., Zapater, M., Alepuz, P.M. et al. 2004. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427, 370–374.
Draper, N.P., Smith, H. 1998. Applied Regression Analysis. Wiley, New York.
Estruch, F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiology Reviews 24, 469–486.
Gasch, A.P., Spellman, P.T., Kao, C.M. et al. 2000. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Molecular Biology of the Cell 11, 4241–4257.
Hahn, J.S., Hu, Z.Z., Thiele, D.J. et al. 2004. Genomewide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24, 5249–56.
Harbison, C.T., Gordon, D.B., Lee, T.I. et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104.
Hocking, R.R. 1976. The analysis and selection of variables in linear regression. Biometrics 32, 1–49.
Hurvich, C.M., Tsai, C.L. 1989. Regression and time series model selection in small samples. Biometrika 76, 297–307.
Iyer, V.R., Horak, C.E., Scafe, C.S. et al. 2001. Genomic binding sites of the yeast cell-cycle transcription factors of SBF and MBF. Nature 409, 533–538.
Jamieson, D.J. 1998. Oxidative Stress Responses of the Yeast Saccharomyces crevisiae. Yeast 14, 1511–1527.
Johansson, R. 1993. System modeling and identification. Prentice-Hall, Englewood Cliffs, New Jersey.
Lee, T.I., Rinaldi, N.J., Robert, F. et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804.
Martínez-Pastor, M.T., Marchler, G., Schüller, C.et al. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15, 2227–2235.
Michael, J.B., Jason, D.L. 2004. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83, 349–360.
Morgan, B.A., Banks, G.R., Toone, W.M., Raitt, D., Kuge, S., Johnston, L.H. 1997. The Skn7 Response Regulator Controls Gene Expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J 16, 1035–1044.
Raitt, D.C., Johnson, A.L., Erkine, A.M., Makino, K., Morgan, B., Gross, D.S., Johnston, L.H. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol Biol Cell 1, 2335–2347.
Ren, B., Robert, F., Wyrick, J.J. et al. 2000. Genomewide location and function of DNA binding proteins. Science 290, 2306–2309.
Rep, M., Reiser, V., Gartner, U. et al. 1999. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol 19, 5474–85.
Rodrigues-Pousada, C.A., Nevitt, T., Menezes, R. et al. 2004. Yeast activator proteins and stress response: an overview. FEBS Letters 567, 80–85.
Ruis, H., Schüller, C. 1995. Stress signaling in yeast. Bioessays 17, 959–965.
Schmitt, A.P., McEntee, K. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of themultistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci 93, 5777–82.
Seber, G.A., Lee, A.J. 2003. Linear Regression Analysis. Wiley-Interscience, New York.
Segal, E., Shapira, M., Regev, A. et al. 2003. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature genetics 34, 166–176.
Sugiyama, K., Izawa, S., Inoue, Y. 2000. The Yap1pdependent induction of glutathione synthesis in heat shock response of Saccharomyces cerevisiae. J Biol Chem 275, 15535–15540.
Uemura, H., Jigami, Y. 1992. Role of GCR2 in transcriptional activation of yeast glycolytic genes. Mol Cell Biol 12, 3834–42.
Wu, W.S., Li, W.H., Chen, B.S. 2006. Computational reconstruction of transcriptional regulatory modules of the yeast cell cycle. BMC Bioinformatics 7, 421–435.
Yamamoto, A., Ueda, J., Yamamoto, N. et al. 2007. Role of Heat Shock Transcription Factor in Saccharomyces cerevisiae Oxidative Stress Response. Eukaryotic Cell 6, 1373–1379.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found online at http://dx.doi.org/10.1007/s12539-009-0018-9
Rights and permissions
About this article
Cite this article
Chen, T., Li, F. & Chen, BS. Cross-talks of sensory transcription networks in response to various environmental stresses. Interdiscip Sci Comput Life Sci 1, 46–54 (2009). https://doi.org/10.1007/s12539-008-0018-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-008-0018-1