Abstract
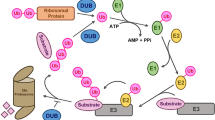
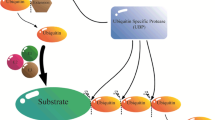
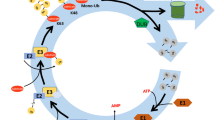
Posttranslational modifications of proteins by small polypeptides including ubiquitination, neddylation (related to ubiquitin (RUB) conjugation), and sumoylation are implicated in plant growth and development, and they regulate protein degradation, location, and interaction with other proteins. Ubiquitination mediates the selective degradation of proteins by the ubiquitin (Ub)/proteasome pathway. The ubiquitin-like protein RUB is conjugated to cullins, which are part of a ubiquitin E3 ligase complex that is involved in auxin hormonal signaling. Sumoylation, by contrast, is known for its involvement in guiding protein interactions related to abiotic and biotic stresses and in the regulation of flowering time. ATG8/ATG12-mediated autophagy influences degradation and recycling of cellular components. Other ubiquitin-like modifiers (ULPs) such as homology to Ub-1, ubiquitin-fold modifier 1, and membrane-anchored Ub-fold are also found in Arabidopsis. ULPs share similar three-dimensional structures and a conjugation system, including E1 activating enzymes, E2 conjugation enzymes, and E3 ligases, as well as proteases for deconjugation and recycling of the tags. However, each of the ULP posttranslational modifications possesses its own specific enzymes and modifies its specific targets selectively. This review discusses recent findings on ubiquitination and ubiquitin-like modifier processes and their roles in the posttranslational modification of proteins in Arabidopsis.


Similar content being viewed by others
References
Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F (2001) Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci 6:463–470
Bassham DC (2007) Plant autophagy—more than a starvation response. Curr Opin Plant Biol 10:587–593
Bassham DC (2009) Function and regulation of macroautophagy in plants. BBA-Mol Cell Res 1793:1397–1403
Bates PW, Vierstra RD (1999) UPL1 and 2, two 405 kDa ubiquitin protein ligases from Arabidopsis thaliana related to the HECT-domain protein family. Plant J 20:183–195
Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280:275–286
Bostick M, Lochhead SR, Honda A, Palmer S, Callis J (2004) Related to ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell 16:2418–2432
Callis J, Carpenter T, Sun CW, Vierstra RD (1995) Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 139:921–939
Capron A, ÖKrész L, Genschik P (2003) First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci 8:83–89
Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua N (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19:2952–2966
Chadwick BP, Kidd T, Sgouros J, Ish-Horowicz D, Frischauf A (1999) Cloning, mapping and expression of UBL3, a novel ubiquitin-like gene. Gene 233:189–195
Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448:666–671
Chosed R, Mukherjee S, Lois LM, Orth K (2006) Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis sumoylation. Biochem J 398:521–529
Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 62:483–493
Colby T, Matthäi A, Boeckelmann A, Stuible H (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142:318–332
de la Cruz NB, Peterson FC, Lytle BL, Volkman BF (2007) Solution structure of a membrane-anchored ubiquitin-fold (MUB) protein from Homo sapiens. Protein Sci 16:1479–1484
del Pozo JC, Estelle M (1999) The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. PNAS 96:15342–15347
del Pozo JC, Timpte C, Tan S, Callis J, Estelle M (1998) The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280:1760–1763
del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 Is required for auxin response. Plant Cell 14:421–433
Deng XW, Caspar T, Quail PH (1991) cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Gene Dev 5:1172–1182
Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M (2003) The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J 22:1762–1770
Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jurgens G, Estelle M (2007) AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J 52:114–123
Dittmar GAG, Wilkinson CRM, Jedrzejewski PT, Finley D (2002) Role of a ubiquitin-like modification in polarized morphogenesis. Science 295:2442–2446
Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277:33105–33114
Dowil R, Lu X, Saracco SA, Vierstra RD, Downes BP (2011) Arabidopsis membrane-anchored ubiquitin-fold (MUB) proteins localize a specific subset of ubiquitin-conjugating (E2) enzymes to the plasma membrane. J Biol Chem 286:14913–14921
Downes BP, Vierstra RD (2005) Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem Soc T 33:393–399
Downes BP, Stupar RM, Gingerich DJ, Vierstra RD (2003) The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35:729–742
Downes BP, Saracco SA, Lee SS, Crowell DN, Vierstra RD (2006) MUBs, a family of ubiquitin-fold proteins that are plasma membrane-anchored by prenylation. J Biol Chem 281:27145–27157
Dreher K, Callis J (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot-London 99:787–822
Faye L, Boulaflous A, Benchabane M, Gomord V, Michaud D (2005) Protein modifications in the plant secretory pathway: current status and practical implications in molecular pharming. Vaccine 23:1770–1778
Fujioka Y, Noda NN, Fujii K, Yoshimoto K, Ohsumi Y, Inagaki F (2008) In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J Biol Chem 283:1921–1928
Gray WM, Hellmann H, Dharmasiri S, Estelle M (2002) Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14:2137–2144
Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282:37298–37302
Harper WE, Burton JL, Solomon MJ (2002) The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev 16:2179–2206
Hermkes R, Fu YF, Nürrenberg K, Budhiraja R, Schmelzer E, Elrouby N, Dohmen RJ, Bachmair A, Coupland G (2011) Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta 233:63–73
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Hotton SK, Callis J (2008) Regulation of cullin RING ligases. Annu Rev Plant Biol 59:467–489
Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, Yang C (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60:666–678
Huffaker RC (1990) Proteolytic activity during senescence of plants. New Phytologist 116:199–231
Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21:2284–2297
Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim W, Oosten MV, Hyun Y, Somers DE, Lee I, Yun D, Bressan RA, Hasegawa PM (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53:530–540
Kang SH, Kim GR, Seong M, Baek SH, Seol JH, Bang OS, Ovaa H, Tatsumi K, Komatsu M, Tanaka K, Chung CH (2007) Two novel ubiquitin-fold modifier 1 (ufm1)-specific proteases, UfSP1 and UfSP2. J Biol Chem 282:5256–5262
Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. PNAS 105:7100–7105
Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22:159–180
Kirschner M (1999) Intracellular proteolysis. Trends Cell Biol 9:M42–M45
Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T, Tanaka K (2004) A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J 23:1977–1986
Kumeta H, Watanabe M, Nakatogawa H, Yamaguchi M, Ogura K, Adachi W, Fujioka Y, Noda N, Ohsumi Y, Inagaki F (2010) The NMR structure of the autophagy-related protein Atg8. J Biomol NMR 47:237–241
Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung D, Vierstra RD (2003) The small ubiquitin-like modifier (sumo) protein modification system in Arabidopsis. Accumulation of SUMO1 and −2 conjugates is increased by stress. J Biol Chem 278:6862–6872
Larsen PB, Cancel JD (2004) A recessive mutation in the RUB1-conjugating enzyme, RCE1, reveals a requirement for RUB modification for control of ethylene biosynthesis and proper induction of basic chitinase and PDF1.2 in Arabidopsis. Plant J 38:626–638
Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, Kim D, Lee SY, Salt DE, Mengiste T, Gong Q, Ma S, Bohnert HJ, Kwak SS, Bressan RA, Hasegawa PM, Yun DJ (2006) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49:79–90
Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364:161–164
Mann M, Jensen ON (2003) Proteomic analysis of post-translational modifications. Nat Biotech 21:255–261
Melchior F (2000) SUMO-nonclassical ubiquitin. Annu Rev Cell Dev Biol 16:591–626
Miura K, Hasegawa PM (2010) Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol 20:223–232
Miura K, Jin JB, Hasegawa PM (2007a) Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10:495–502
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Hasegawa PM (2007b) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19:1403–1414
Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. PNAS 106:5418–5423
Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16:3181–3195
Ohsumi Y, Mizushima N (2004) Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol 15:231–236
Phillips AR, Suttangkakul A, Vierstra RD (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178:1339–1353
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503
Ramelot TA, Cort JR, Yee AA, Semesi A, Edwards AM, Arrowsmith CH, Kennedy MA (2003) Solution structure of the yeast ubiquitin-like modifier protein Hub1. J Struct Funct Genomics 4:25–30
Rao-Naik C, dela Cruz W, Laplaza JM, Tan S, Callis J, Fisher AJ (1998) The rub family of ubiquitin-like proteins. J Biol Chem 273:34976–34982
Refy AE, Perazza D, Zekraoui L, Valay J, Bechtold N, Brown S, Hülskamp M, Herzog M, Bonneville J (2003) The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol Genet Genomics 270:403–414
Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007) Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145:119–134
Sasakawa H, Sakata E, Yamaguchi Y, Komatsu M, Tatsumi K, Kominami E, Tanaka K, Kato K (2006) Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1. Biochem Biophy Res Co 343:21–26
Serino G, Deng X (2003) The COP9 signalosome: regulating plant development through the control of proteolysis. Annu Rev Plant Biol 54:165–182
Smalle J, Vierstra RD (2004) The ubiquitin 26s proteasome proteolytic pathway. Annu Rev Plant Biol 55:555–590
Stone SL, Anderson EM, Mullen RT, Goring DR (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible brassica pollen. Plant Cell 15:885–898
Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F (2005) The crystal structure of plant ATG12 and its biological implication in autophagy. Autophage 1:119–126
Tatsumi K, Sou Y, Tada N, Nakamura E, Iemura S, Natsume T, Kang SH, Chung CH, Kasahara M, Kominami E, Yamamoto M, Tanaka K, Komatsu M (2010) A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem 285:5417–5427
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448:661–665
Thomann A, Dieterle M, Genschik P (2005) Plant CULLIN-based E3s: phytohormones come first. FEBS Lett 579:3239–3245
Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8:165–173
Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138:2097–2110
van den Burg HA, Kini RK, Schuurink RC, Takken FLW (2010) Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 22:1998–2016
Vierstra RD (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10:385–397
Vijay-kumar S, Bugg CE, Cook WJ (1987) Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol 194:531–544
Vinarov DA, Lytle BL, Peterson FC, Tyler EM, Volkman BF, Markley JL (2004) Cell-free protein production and labeling protocol for NMR-based structural proteomics. Nat Meth 1:149–153
Wang KL, Yoshida H, Lurin C, Ecker JR (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428:945–950
Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19:261–286
Wilkinson CRM, Dittmar GA, Ohi MD, Uetz P, Jones N, Finley D (2004) Ubiquitin-like protein Hub1 is required for pre-mRNA splicing and localization of an essential splicing factor in fission yeast. Curr Biol 14:2283–2288
Wold F (1981) In vivo chemical modification of proteins (post-translational modification). Annu Rev Biochem 50:783–814
Woodward AW, Ratzel SE, Woodward EE, Shamoo Y, Bartel B (2007) Mutation of E1-conjugating enzyme-related1 decreases related to ubiquitin conjugation and alters auxin response and development. Plant Physiol 144:976–987
Xie Q, Guo H, Dallman G, Fang S, Weissman AM, Chua N (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419:167–170
Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD (2000) The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124:1828–1843
Yashiroda H, Tanaka K (2004) Hub1 is an essential ubiquitin-like protein without functioning as a typical modifier in fission yeast. Genes Cells 9:1189–1197
Yee D, Goring DR (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60:1109–1121
Yi C, Deng XW (2005) COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15:618–625
Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun D, Bressan RA, Hasegawa PM (2006) SIZ1 small ubiquitin-like modifier e3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142:1548–1558
Yoshida H, Nagata M, Saito K, Wang K, Ecker J (2005) Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biology 5:14
Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16:2967–2983
Ytterberg AJ, Jensen ON (2010) Modification-specific proteomics in plant biology. J Proteomics 73:2249–2266
Acknowledgments
This work was supported by grants from the World Class University Program (R32-10148), the Next-Generation BioGreen 21 Program (SSAC, PJ008025), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, H.J., Park, H.C., Lee, S.Y. et al. Ubiquitin and Ubiquitin-like Modifiers in Plants. J. Plant Biol. 54, 275–285 (2011). https://doi.org/10.1007/s12374-011-9168-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-011-9168-5