Abstract
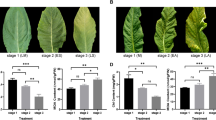
Premature senescence of leaves can critically influence tomato yield and quality. In this study, the leaf premature senescence mutant MT318 was a spontaneous mutant and was controlled by a single recessive nuclear gene. The maximum photochemical efficiency (Fv/Fm), superoxide dismutase (SOD), and chlorophyll content in the leaves of mutant MT318 gradually decreased, while malondialdehyde (MDA) content significantly increased. Under the level 2 category, Gene Ontology (GO) enrichment analysis indicated that 45 terms were enriched, comprising 22 in biological process, 12 in cellular component, and 11 in molecular function. Genes are mainly involved in the metabolic processes (696 differentially expressed genes, DEGs), cellular processes (573 DEGs), single-organism processes (503 DEGs), and catalytic activity (675 DEGs). Kyoto Encyclopedia of Genes and Genomes pathway analysis demonstrated that the 4 pathways with the largest number of genes were biosynthesis of secondary metabolites, plant-pathogen interaction, plant hormone signal transduction, and MAPK signaling pathway-plant. The ‘plant hormone signal transduction’ pathway was the most significantly enriched at the T2 stage. Pearson correlation analysis showed that the auxin regulatory pathway and SA signal transduction pathway may play important roles. These results not only lay the foundation for the further cloning and functional analysis of the MT318 premature senescence gene but also provide a reference for the study of tomato leaf senescence.









Similar content being viewed by others
Data availability
The transcriptome data used in this study have been submitted to the NCBI database under accession number PRJNA813642.
References
Astrid W, Sarah P, Maclean JA, Nathalie P (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J Exp Bot 2:391–399. https://doi.org/10.1093/jxb/eri279
Balazadeh S, Riano-Pachon DM, Mueller-Roeber B (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol 10(s1):63–75. https://doi.org/10.1111/j.1438-8677.2008.00088.x
Bartrina I, Otto E, Strnad M, Schmülling WT (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23(1):69–80. https://doi.org/10.2307/41433804
Bouzayen M (2012) Genome-Wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53(4):659–672. https://doi.org/10.1093/pcp/pcs022
Cang J, Yu LF, Wang YY, Zhang D, Hao ZB, Hu BZ (2007) Agronomic and biochemical properties chlorophyll deletion mutant HS 821of soybean. J Nucl Agric Sci 21(001):9–12. https://doi.org/10.3969/j.issn.1000-8551.2007.01.003
Cao L, Wang H, Sun SJ, Feng Y, Li XJ, Min DH (2008) Genetic analysis of a novel aurea mutant in wheat. Hereditas 30(12):1603–1607. https://doi.org/10.3724/SP.J.1005.2008.01603
Cha KW, Lee YJ, Koh HJ, Lee BM, Nam YW, Paek NC (2002) Isolation, characterization, and mapping of the stay green mutant in rice. Theor Appl Genet 104(4):526–532. https://doi.org/10.1007/s001220100750
Chen S, Zhou Y, Chen Y, Jia G (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chen Y, Feng P, Tang B, HuZ CG (2022) The AP2/ERF transcription factor SlERF.F5 functions in leaf senescence in tomato. Plant Cell Rep 41:1181–1195. https://doi.org/10.1007/s00299-022-02846-1
Cheng ZL, Wang YQ, Zhu YN, Tang JY, Hu B (2014) OsNAP connects abscisic acid and leafsenescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. PNAS 111(7):10013–10018. https://doi.org/10.1073/pnas.1321568111
Chinnusamy V, Jagendorf A, Zhu J (2005) Understanding and improving salt tolerance in plants. Crop Sci 45(2):437–448. https://doi.org/10.2135/cropsci2005.0437
Cui LM, Song LH, Huang ZQ, Gao JC, Guo YM, Du YC, Wang XX (2017) Genetic mapping and physiological characteristics of tomato yellowing gene netted viresce (NV). China Veg 7:29–36
Fan YY, Liu YM, Li ZS, Fang ZY, Yang LM, Zhuang M, Zhang YY, Sun PT (2015) Analysis of the expression of chlorophyll degrading genes during senescence of broccoli. Acta Hortic Sin 42(7):1338–1346. https://doi.org/10.16420/j.issn.0513-353x.2015-0155
Fukao T, Bailey-Serres YJ (2012) The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol 160(4):1795–1807. https://doi.org/10.2307/41812026
Gao Q, Yang Z, Zhou Y, Yin Z, Qiu J, Liang G, Xu C (2012) Characterization of an Abc1 kinase family gene OsABC1-2 conferring enhanced tolerance to dark-induced stress in rice. Gene 498(2):155–163. https://doi.org/10.1016/j.gene.2012.02.017
Gonzalez GM, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Rodriguez K (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24(6):2483–2496. https://doi.org/10.1105/tpc.112.098574
Guan XX, Sui CL, Li JH, Xu T, Hu HJ, Wu YN (2018) Effects of tomato SlARF2 gene on leaf senescence. Genomics Appl Biol 37(1):393–399
Han XX, Han JX, Jiang J (2015) Screening the reference genes for the studies of Quantitative real-time RT-PCR in tomato under Abiotic stress. Mol Plant Breed 13(4):822–831. https://doi.org/10.13271/j.mpb.013.000822
Hansson A, Kannangara CG, Wettstein DV, Hansson M (1999) Molecular basis for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of magnesium chelatase. PNAS 96(4):1744–1749. https://doi.org/10.1073/pnas.96.4.1744
Huang XQ, Wang PR, Zhao HX, Deng XJ (2007) Genetic analysis and molecular mapping of a novel chlorophyll-deficit mutant gene in rice. Chin J Rice Sci 21(4):355–359. https://doi.org/10.3321/j.issn:1001-7216.2007.04.005
Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Fan C, Wu G (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52(2):197–209. https://doi.org/10.1111/j.1365-313X.2007.03221.x
Jiang J, Wang YL, Zhao LP, Zhou R, Li YR, Zhao TM, Yu WG (2017) Selection of tomato reference genes for qRT-PCR. Jiangsu J Agric Sci 33(2):389–396. https://doi.org/10.3969/j.issn.1000-4440.2017.02.024
Jiang B (2020) The mechanism of SlPP2C regulating tomato senescence. Dissertation, Shenyang Agricultural University
Jibran R, Hunter D, Dijkwel P (2013) Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol Biol 82(6):547–561. https://doi.org/10.1007/s11103-013-0043-2
Khan M, Rozhon W, Poppenberger B (2013) The Role of hormones in the aging of plants—a mini-review. Gerontology 60(1):49–55. https://doi.org/10.1159/000354334
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. https://doi.org/10.1038/nmeth.3317
Kong Z, Li M, Xue XY, Yang W (2006) A novel nuclear-Localized CCCH-Type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141(4):1376–1388. https://doi.org/10.1104/pp.106.082941
Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M (2007) Rice Non-Yellow coloring1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19(4):1362–1375. https://doi.org/10.1105/tpc.106.042911
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. https://doi.org/10.1038/nmeth.1923
Lee S, Seo PJ, Lee HU, Park CO (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought induced leaf senescence in Arabidopsis. Plant J 70(5):831–844. https://doi.org/10.1111/j.1365-313X.2012.04932.x
Li HS (2000) Plant physiological and biochemical experiment principles and techniques. Beijing Higher Education Press Version 3, pp 274–276
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In: Methods in enzymology, vol 148. Academic Press, pp 350–382. https://doi.org/10.1016/0076-6879(87)48036-1
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136. https://doi.org/10.1146/annurev.arplant.57.032905.105316
Lin A, Wang Y, Tang J, Xue P, Li C, Liu L, Hu B, Yang F, Loake GJ, Chu C (2011) Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol 158(1):451–464. https://doi.org/10.1104/pp.111.184531
Liu L, Yong Z, Zhou G, Ye R, Zhao L, Li X, Lin Y (2008) Identification of early senescence-associated genes in rice flag leaves. Plant Mol Biol 67(1–2):37–55. https://doi.org/10.1007/s11103-008-9300-1
Liu FZ, Zhang Y, Yang JK, Chen YH, Shu JS, Li SP, Chen LL (2020) Characterization and genetic analysis of a yellowing mutant of eggplant leaf color. Acta Hortic Sin 47(12):2340–2348. https://doi.org/10.16420/j.issn.0513-353x.2020-0829
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Luo PG, Ren ZL (2006) Wheat leaf chlorosis controlled by a single recessive gene. J Plant Physiol Mol Biol 32(3):330. https://doi.org/10.3321/j.issn:1671-3877.2006.03.011
Ma XM, Zhang YJ, Tureckova V, Xue GP, Fernie A (2018) The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol 177:1286–1302. https://doi.org/10.1104/pp.18.00292
Mallory AC (2005) MicroRNA-Directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17(5):1360–1375. https://doi.org/10.1105/tpc.105.031716
Mason MG, Mathews DE, Argyros DA, Maxwell BB, Schaller GE (2005) Multiple Type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17(11):3007–3018. https://doi.org/10.1105/tpc.105.035451
Miao Y, Geng MY, ShenPF CXH, Li YJ (2018) Effect of post-silking drought stress on the expression profiles of genes involved in carbon and nitrogen metabolism during leaf senescence in maize (Zea mays L.). Plant Physiol Biochem 135:304–309. https://doi.org/10.1016/j.plaphy.2018.12.025
Moschen S, Fernández P, Luoni SB, Paniego N, Aguirrezábal L, Dosio G, Heinz RA (2012) Sunflower (Helianthus annuus L.) leaf senescence analysis by a physiological and molecular approach. In: 18th Sunflower Int Conf.
Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61(2):290–299. https://doi.org/10.1111/j.1365-313X.2009.04054.x
Nothnagel T, Straka P (2003) Inheritance and mapping of a yellow leaf mutant of carrot (Daucus carota). Plant Breed 122(4):339–342. https://doi.org/10.1046/j.1439-0523.2003.00884.x
Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19(1):118–130. https://doi.org/10.1105/tpc.106.047761
Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19:1649–1664. https://doi.org/10.2307/20077046
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290. https://doi.org/10.1038/nbt.3122
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11(9):1650–1667. https://doi.org/10.1038/nprot.2016.095
Sakakibara H (2006) CYTOKININS: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57(1):431–449. https://doi.org/10.1146/annurev.arplant.57.032905.105231
Sakuraba Y, Rahman ML, Cho SH, Kim YS, Koh HJ, Yoo SC, Paek NC (2013) The rice faded green leaf locus encodes protochlorophyllide oxidoreductaseB and is essential for chlorophyll synthesis under high light conditions. Plant J 74(1):122–133. https://doi.org/10.1111/tpj.12110
Shen HL, Cheng JS, Han QX (2006) Studies on inheritance and phenotype of the yellow mutant of carrot. Acta Hortic Sin 33(4):856–858. https://doi.org/10.3321/j.issn:0513-353X.2006.04.035
Shu Z, Zhang XS, Chen J, Chen GY, Xu DQ (2010) The simplification of chlorophyll content measurement. Plant Physiol J 46(4):399–402
Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7(11):847–859. https://doi.org/10.1038/nrm2020
Tiwari SB (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15(2):533. https://doi.org/10.1105/tpc.008417
To J, Haberer G, Ferreira FJ, Deruère J, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16(3):658–671. https://doi.org/10.1105/tpc.018978
Wang S, Tiwari SB, Hagen G, Guilfoyle TJ (2005) AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant arabidopsis leaf mesophyll protoplasts. Plant Cell 17(7):1979–1993. https://doi.org/10.1109/38.403820
Wang W, Xu YJ, Wan ZJ (2011) Cloning and expression analsis of key genes PPH and PAO for chlorophyll degradation in cucumber. Acta Hortic Sin 38(6):1104–1110
Wang ZX, Tang ZS (1996) Genetic analysis of yellow leaf colour mutant in Barley, Southwest China. J Agric Sci (S1):180–182
Woodward AW, Bartel B (2005) A receptor for auxin. Plant Cell 17(9):2425–2429. https://doi.org/10.2307/4130926
Xu N, Xu JM, Jiang LH, Rao YC (2017) Advances in understanding leaf premature senescence and its molecular mechanism in rice. Chin Bull Bot 52(1):102–112. https://doi.org/10.11983/CBB16222
Yamatani H, SatoY MY, Kato Y, Morita R, Fukunaga K, Nagamura Y, Nishimura M, Sakamoto W, Tanaka A (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll—protein complexes during leaf senescence. Plant J 74(4):652–662. https://doi.org/10.1111/tpj.12154
Yang Y, Tang N, Li ZG (2014) The role of SlSnRK2s expression in promoting the leaf senecence in tomato. Chin J Trop Crops 35(5):950–956. https://doi.org/10.3969/j.issn.1000-2561.2014.05.020
Yang XM, Wu XL, Liu YF, Li TL, Qi MF (2018) Analysis of chlorophyll and photosynthesis of a tomato chlorophyll-deficient nutant induced by EMS. Chin J Appl Ecol 29(6):1983–1989. https://doi.org/10.13287/j.1001-9332.201806.021
Yang Y (2013) Response of tomato SlPYL/SlPP2C/SlSnRK2 gene family to ABA and osmotic stress and functional study of SlSnRK2s gene. Dissertation, Chongqing University
Ys A, Dk A, Ysk A, Sh B, Ncp A (2014) Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett 588(21):3830–3837. https://doi.org/10.1016/j.febslet.2014.09.018
Zentgraf U, Laun T, Ying M (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol 89(2–3):133–137. https://doi.org/10.1016/j.ejcb.2009.10.014
Zhang K, Novak O, Wei Z, Gou M, Zhang X, Yu Y, Yang H, Cai Y, Strnad M, Liu CJ (2014) Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat Commun 5:327. https://doi.org/10.1038/ncomms4274
Zhang TY, Zhou CL, Liu X, Sun AL, Cao PH, Nguyen T, Tian YL, Zhai HQ, Jiang L (2017) Phenotypic analysis and gene mapping of a thermo sensitive yellowing mutant in rice. Acta Agron Sin 43(10):1426–1433. https://doi.org/10.3724/SP.J.1006.2017.01426
Zhang Q, Xia C, Zhang L, Dong C, Xu L, Kong X (2018) Transcriptome analysis of a premature leaf senescence mutant of common wheat (Triticum aestivum L.). Int J Mol Sci 19(3):782. https://doi.org/10.3390/ijms19030782
Zhao L, Yan X, Wu XY, Schippers J, Jing HC (2018) Phenotypic analysis and molecular markers of leaf senescence. Methods Mol Biol 1744:35–48. https://doi.org/10.1007/978-1-4939-7672-0_3
Acknowledgements
This study was supported by theTianjin Science and Technology Planning Project (No. 21ZYCGN00420). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
JF designed the experiments and wrote this paper. HM,SL, CS,SH, KW, HZ conducted the experiments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jin, F., Hua, M., Song, L. et al. Transcriptome analysis of gene expression in the tomato leaf premature senescence mutant. Physiol Mol Biol Plants 28, 1501–1513 (2022). https://doi.org/10.1007/s12298-022-01223-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-022-01223-2