Abstract
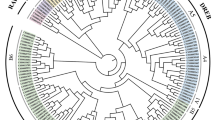
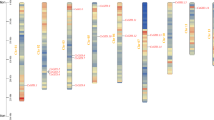
Phospholipase D (PLD) (EC 3.1.4.4) plays important roles in plants growth, development, and response to environmental stresses. Tea plant (Camellia sinensis) is the most important non-alcoholic beverage in the world with health benefits, but tea production decreases in response to environmental stresses such as cold and drought. Therefore, a genome-wide analysis of the C. sinensis PLD gene family (CsPLDs) was carried out. In the current study, identification, evolutionary relationship, duplication, selection pressure, gene structure, promoter analysis, transcript-targeted miRNA, and simple sequence repeat markers prediction, RNA-seq data analysis, and three-dimensional structure of the CsPLDs have been investigated using bioinformatics tools. 15 PLDs were identified from the tea genome which belongs to five groups, including CsPLDα, CsPLDβ, CsPLDδ, CsPLDε, and CsPLDζ. Both segmental and tandem duplications have occurred in the CsPLD gene family. Ka/Ks ratio for the most duplicated pair genes was less than 1 which implies negative selection to conserve their function during the tea evolution. 68 cis-elements have been found in CsPLDs indicating the contribution of these genes in response to environmental stresses. Likewise, 72 SSR loci and 96 miRNA molecules in 14 and 15 CsPLDs have been detected. According to RNA-seq data, the highest expression in all tissues under various abiotic stresses was related to CsPLDα1. Besides, a three-dimensional structure of the CsPLDα1 was evaluated to better understand its biological activity. This research provides comprehensive information that could be useful in future studies to develop stress-tolerant tea.








Similar content being viewed by others
References
Angelini J, Vosolsobě S, Skůpa P, Ho AYY, Bellinvia E, Valentová O, Marc J (2018) Phospholipase Dδ assists to cortical microtubule recovery after salt stress. Protoplasma 255:1195–1204
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Bargmann BO et al (2009) Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol 50:78–89
Boualem A et al (2008) MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J 54:876–887
Bowling FZ, Salazar CM, Bell JA, Huq TS, Frohman MA, Airola MV (2020) Crystal structure of human PLD1 provides insight into activation by PI (4, 5) P 2 and RhoA. Nat Chem Biol 16:400–407
Chen L, Cao B, Han N, Tao Y, Zhou SF, Li WC, Fu FL (2017) Phospholipase D family and its expression in response to abiotic stress in maize. Plant Growth Regul 81:197–207
Chen C, Chen H, He Y, Xia R (2018) TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv:289660
Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46:W49–W54
Du D, Cheng T, Pan H, Yang W, Wang J, Zhang Q (2013) Genome-wide identification, molecular evolution and expression analyses of the phospholipase D gene family in three Rosaceae species. Sci Horticult 153:13–21
Eliáš M, Potocký M, Cvrčková F, Žárský V (2002) Molecular diversity of phospholipase D in angiosperms. BMC Genomics 3:2
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Finn RD et al (2013) Pfam: the protein families database. Nucleic Acids Res:gkt1223
Goodstein DM et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186
Gull A, Lone AA, Wani NUI (2019) Biotic and abiotic stresses in plants. Abiotic Biotic Stress Plants, pp 1–19
Hajiahmadi Z, Abedi A, Wei H, Sun W, Ruan H, Zhuge Q, Movahedi A (2020) Identification, evolution, expression, and docking studies of fatty acid desaturase genes in wheat (Triticum aestivum L.). BMC genom 21:1–20
Hong Y, Pan X, Welti R, Wang X (2008) Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20:803–816
Hong Y, Zhang W, Wang X (2010) Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant Cell Environ 33:627–635
Hong K, Zhang L, Zhan R, Huang B, Song K, Jia Z (2017) Identification and characterization of phospholipase d genes putatively involved in internal browning of pineapple during postharvest storage. Front Plant Sci 8:913
Howe KL et al (2019) Ensembl Genomes 2020—enabling non-vertebrate genomic research. Nucleic Acids Res 48:D689–D695
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics:btu817
Jha A, Shankar R (2014) MiRNAting control of DNA methylation. J Biosci 39:365–380
Ji T et al (2017) Overexpression of cucumber phospholipase D alpha gene (CsPLDα) in tobacco enhanced salinity stress tolerance by regulating Na+–K+ balance and lipid peroxidation. Front Plant Sci 8:499
Johnson DA, Thomas MA (2007) The monosaccharide transporter gene family in Arabidopsis and rice: a history of duplications, adaptive evolution, and functional divergence. Molecular Biol Evolut 24:2412–2423
Katagiri T, Takahashi S, Shinozaki K (2001) Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J 26:595–605
Kumar S, Stecher G, Tamura K (2016a) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biol Evolut 33:1870–1874
Kumar S, Stecher G, Tamura K (2016b) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biol Evolut:msw054
Laskowski RA, Chistyakov VV, Thornton JM (2005) PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res 33:D266–D268
Lein W, Saalbach G (2001) Cloning and direct G-protein regulation of phospholipase D from tobacco. Biochim Biophys Acta (BBA)-Molecular Cell Biol Lipids 1530:172–183
Leiros I, McSweeney S, Hough E (2004) The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J Molecular Biol 339:805–820
Lescot M et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li W, Li M, Zhang W, Welti R, Wang X (2004) The plasma membrane–bound phospholipase Dδ enhances freezing tolerance in Arabidopsis thaliana. Nature Biotechnol 22:427–433
Li G, Lin F, Xue H-W (2007) Genome-wide analysis of the phospholipase D family in Oryza sativa and functional characterization of PLDβ1 in seed germination. Cell Res 17:881–894
Li W, Song T, Wallrad L, Kudla J, Wang X, Zhang W (2019) Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nature Plants 5:1012–1021
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu Q, Zhang C, Yang Y, Hu X (2010) Genome-wide and molecular evolution analyses of the phospholipase D gene family in Poplar and Grape. BMC Plant Biol 10:117
Long M, Deutsch M (1999) Association of intron phases with conservation at splice site sequences and evolution of spliceosomal introns. Molecular Biol Evolut 16:1528–1534
Lovell SC et al (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct Funct Bioinformatics 50:437–450
Lu S, Fadlalla T, Tang S, Li L, Ali U, Li Q, Guo L (2019) Genome-wide analysis of Phospholipase D gene family and profiling of phospholipids under abiotic stresses in Brassica napus. Plant Cell Physiol 60:1556–1566
Margutti MP, Reyna M, Meringer MV, Racagni GE, Villasuso AL (2017) Lipid signalling mediated by PLD/PA modulates proline and H2O2 levels in barley seedlings exposed to short-and long-term chilling stress. Plant Physiol Biochem 113:149–160
Meng Y, Shao C, Ma X, Wang H, Chen M (2012) Expression-based functional investigation of the organ-specific microRNAs in Arabidopsis. PLoS ONE 7:e50870
Mukhopadhyay M, Mondal TK, Chand PK (2016) Biotechnological advances in tea (Camellia sinensis [L.] O. Kuntze): a review. Plant Cell Rep 35:255–287
Othman AB et al (2017) Phospholipases Dζ1 and Dζ2 have distinct roles in growth and antioxidant systems in Arabidopsis thaliana responding to salt stress. Planta 246:721–735
Qin C, Wang X (2002) The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol 128:1057–1068
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protocols 5:725
Ruelland E, Kravets V, Derevyanchuk M, Martinec J, Zachowski A, Pokotylo I (2015) Role of phospholipid signalling in plant environmental responses. Environ Exp Bot 114:129–143
Sagar S, Biswas DK, Chandrasekar R, Singh A (2021) Genome-wide identification, structure analysis and expression profiling of phospholipases D under hormone and abiotic stress treatment in chickpea (Cicer arietinum). Int J Biol Macromol 169:264–273
Sang Y, Zheng S, Li W, Huang B, Wang X (2001) Regulation of plant water loss by manipulating the expression of phospholipase Dα. Plant J 28:135–144
Shannon P et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Shi J et al (2015) Transcriptional responses and flavor volatiles biosynthesis in methyl jasmonate-treated tea leaves. BMC Plant Biol 15:1–20
Singh A, Pandey A, Baranwal V, Kapoor S, Pandey GK (2012) Comprehensive expression analysis of rice phospholipase D gene family during abiotic stresses and development. Plant Signal Behav 7:847–855
Song Y et al (2012) Cloning and characterization of a cucumber phospholipase D gene in response to excess nitrate and other abiotic stresses. Sci Horticult 135:128–136
Tang K, Dong C, Liu J (2016) Genome-wide analysis and expression profiling of the phospholipase D gene family in Gossypium arboreum. Sci China Life Sci 59:130–141
Tang K, Dong C-J, Liu J-Y (2016) Genome-wide comparative analysis of the phospholipase D gene families among allotetraploid cotton and its diploid progenitors. PLoS ONE 11:e0156281
Tiwari K, Paliyath G (2011) Cloning, expression and functional characterization of the C2 domain from tomato phospholipase Dα. Plant Physiol Biochem 49:18–32
Upadhyaya H, Panda SK (2013) Abiotic stress responses in tea [Camellia sinensis L (O) Kuntze]: an overview. Rev Agricult Sci 1:1–10
Wan S, Li M, Ma F, Yuan J, Liu Z, Zheng W, Zhan J (2019) Genome-wide identification of phospholipase D (PLD) gene family and their responses to low-temperature stress in peach. In: AIP Conference Proceedings, vol 1. AIP Publishing LLC, p 020011
Wang X (2005) Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol 139:566–573
Wang X, Xu L, Zheng L (1994) Cloning and expression of phosphatidylcholine-hydrolyzing phospholipase D from Ricinus communis L. J Biol Chem 269:20312–20317
Wang X-C et al (2013) Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 14:1–15
Wang Z, Qiao Y, Zhang J, Shi W, Zhang J (2017) Genome wide identification of microRNAs involved in fatty acid and lipid metabolism of Brassica napus by small RNA and degradome sequencing. Gene 619:61–70
Wang J, Xu M, Li Z, Ye Y, Rong H, Xu L-a (2018) Tamarix microRNA profiling reveals new insight into salt tolerance. Forests 9:180
Wang P et al (2019) Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED2 auxin efflux transporter in response to salt stress. Plant Cell 31:250–271
Wei K, Pan S, Li Y (2016) Functional characterization of Maize C 2 H 2 Zinc-Finger Gene Family. Plant Mol Biol Report 34:761–776
Wei C et al (2018) Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci 115:E4151–E4158
Wu C et al (2019) The class III peroxidase (pod) gene family in cassava: identification, phylogeny, duplication, and expression. Int J Molecular Sci 20:2730
Xia EH et al (2019) Tea plant information archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol J 17:1938–1953
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101:2525–2534
Xue T, Liu Z, Dai X, Xiang F (2017) Primary root growth in Arabidopsis thaliana is inhibited by the miR159 mediated repression of MYB33, MYB65 and MYB101. Plant Sci 262:182–189
Yao Y, Li J, Lin Y, Zhou J, Zhang P, Xu Y (2020) Structural insights into phospholipase D function. Progress Lipid Res, 101070
You FM et al (2008) BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9:253
Zhang W, Wang C, Qin C, Wood T, Olafsdottir G, Welti R, Wang X (2003) The oleate-stimulated phospholipase D, PLDδ, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15:2285–2295
Zhang Q, Lin F, Mao T, Nie J, Yan M, Yuan M, Zhang W (2012) Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24:4555–4576
Zhang Q, Cai M, Yu X, Wang L, Guo C, Ming R, Zhang J (2017) Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genet Genomes 13:1–17
Zhang B, Wang Y, Liu J-Y (2018) Genome-wide identification and characterization of phospholipase C gene family in cotton (Gossypium spp.). Sci China Life Sci 61:88–99
Zhao J, Zhou D, Zhang Q, Zhang W (2012) Genomic analysis of phospholipase D family and characterization of GmPLDαs in soybean (Glycine max). J Plant Res 125:569–578
Zhou L et al (2014) Exogenous abscisic acid significantly affects proteome in tea plant (Camellia sinensis) exposed to drought stress. Horticult Res 1:1–9
Zhu Z, Miao Y, Guo Q, Zhu Y, Yang X, Sun Y (2016) Identification of miRNAs involved in stolon formation in Tulipa edulis by high-throughput sequencing. Front Plant Sci 7:852
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest, and the manuscript has been approved by all authors for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Roshan, N.M., Ashouri, M. & Sadeghi, S.M. Identification, evolution, expression analysis of phospholipase D (PLD) gene family in tea (Camellia sinensis). Physiol Mol Biol Plants 27, 1219–1232 (2021). https://doi.org/10.1007/s12298-021-01007-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-01007-0