Abstract
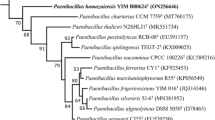
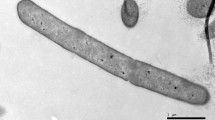
A Gram-stain-positive, rod-shaped, non-endospore-forming motile by means of peritrichous flagella, facultatively anaerobic bacterium designated TI45-13arT was isolated from Nuruk, a Korean traditional Makgeolli fermentation starter. It grew at 4–35°C (optimum, 28–30°C), pH 5.0–9.0 (optimum, pH 7.0) and NaCl concentrations up to 5% (w/v). Phylogenetic trees generated using 16S rRNA gene sequences revealed that strain TI45-13arT belonged to the genus Paenibacillus and showed the highest sequence similarities with Paenibacillus kyungheensis DCY88T (98.5%), Paenibacillus hordei RH-N24T (98.4%) and Paenibacillus nicotianae YIM h-19T (98.1%). The major fatty acid was anteiso-C15:0. The DNA G+C content was 39.0 mol%, and MK-7 was the predominant isoprenoid quinone. The polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, three unidentified glycolipids, and one unidentified aminoglycolipid. The cell-wall peptidoglycan contained meso-diaminopimelic acid. On the basis of polyphasic taxonomy study, it was suggested that strain TI45-13arT represents a novel species within the genus Paenibacillus for which the name Paenibacillus nuruki sp. nov. is proposed. The type strain was TI45-13arT (= KACC 18728T = NBRC 112013T).
Similar content being viewed by others
References
Ash, C., Priest, F.G., and Collins, M.D. 1993. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie van Leeuwenhoek 64, 253–260.
Chin, C.S., Alexander, D.H., Marks, P., Klammer, A.A., Drake, J., Heiner, C., Clum, A., Copeland, A., Huddleston, J., Eichler, E.E., et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569.
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D.R., da Costa, M.S., Rooney, A.P., Yi, H., Xu, X.W., De Meyer, S., et al. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466.
de Costa, M.S., Albuquerque, L., Nobre, M.F., and Wait, R. 2011. The identification of polar lipids in prokaryotes. Methods Microbiol. 38, 101–129.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Fitch, W.M. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20, 406–416.
Kim, J.M., Lee, S.H., Lee, S.H., Choi, E.J., and Jeon, C.O. 2013. Paenibacillus hordei sp. nov., isolated from naked barley in Korea. Antonie van Leeuwenhoek 103, 3–9.
Kim, M., Oh, H.S., Park, S.C., and Chun, J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351.
Lagesen, K., Hallin, P.F., Rodland, E., Stærfeldt, H.H., Rognes, T., and Ussery, D.W. 2007. RNAmmer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 35, 3100–3108.
Lane, D.J. 1991. 16S/23S rRNA sequencing, pp. 115–175. In Stackebrandt. E. and Goodfellow, M. (eds.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, FL, USA.
Li, Q.Q., Zhou, X.K., Dang, L.Z., Cheng, J., Hozzein, W.N., Liu, M.J., Hu, Q., Li, W.J., and Duan, Y.Q. 2014. Paenibacillus nicotianae sp. nov., isolated from a tobacco sample. Antonie van Leeuwenhoek 106, 199–205.
Logan, N.A., Berge, O., Bishop, A.H., Busse, H.J., De Vos, P., Fritze, D., Heyndrickx, M., Kämpfer, P., Rabinovitch, L., Salkinoja-Salonen, M.S., et al. 2009. Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int. J. Syst. Evol. Microbiol. 59, 2114–2121.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Pednekar, P.B., Jain, R., Thakur, N.L., and Mahajan, G.B. 2010. Isolation of multi-drug resistant Paenibacillus sp. from fertile soil: an imminent menace of spreading resistance. J. Life Sci. 4, 7–12.
Pruesse, E., Peplies, J., and Glöckner, F.O. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829.
Saha, P., Krishnamurthi, S., Bhattacharya, A., Sharma, R., and Chakrabarti, T. 2010. Fontibacillus aquaticus gen. nov., sp. nov., isolated from a warm spring. Int. J. Syst. Evol. Microbiol. 60, 422–428.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. Microbial ID Inc., Newark, DE, USA.
Schaeffer, A.B. and Fulton, M.D. 1933. A simplified method of staining endospores. Science 77, 194.
Shida, O., Takagi, H., Kadowaki, K., Nakamura, L.K., and Komagata, K. 1997. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int. J. Syst. Evol. Microbiol. 47, 289–298.
Shin, S.K., Kim, E., and Yi, H. 2018. Paenibacillus crassostreae sp. nov., isolated from the pacific oyster Crassostrea gigas. Int. J. Syst. Evol. Microbiol. 68, 58–63.
Siddiqi, M.Z., Siddiqi, M.H., Im, W.T., Kim, Y.J., and Yang, D.C. 2015. Paenibacillus kyungheensis sp. nov., isolated from flowers of Magnolia. Int. J. Syst. Evol. Microbiol. 65, 3959–3964.
Staneck, J.L. and Roberts, G.D. 1974. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729.
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E.P., Zaslavsky, L., Lomsadze, A., Pruitt, K.D., Borodovsky, M., and Ostell, J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617.
Acknowledgements
This study was carried out with the support (Project no. PJ-011248) of the National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
The GenBank number for the 16S rRNA gene sequence and the genome sequence of strain TI45-13arT are KY419705 and MDER00000000, respectively.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Kim, SJ., Cho, H., Ahn, JH. et al. Paenibacillus nuruki sp. nov., isolated from Nuruk, a Korean fermentation starter. J Microbiol. 57, 836–841 (2019). https://doi.org/10.1007/s12275-019-9118-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-019-9118-3