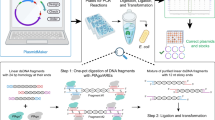
Abstract
Construction of plasmids has been occupying a significant fraction of laboratory work in most fields of experimental biology. Tremendous effort was made to improve the traditional method for constructing plasmids, in which DNA fragments digested with restriction enzymes were ligated. However, the traditional method remained to be a standard protocol more than 40 years. At last, several recent inventions are rapidly and completely replacing the traditional method, because they are far quicker with less cost, and requiring less material. We here introduce three such methods that cover up most of the cases. Moreover, they are complementary with each other. Our lab protocols are provided for “no strain, no pain” construction of plasmids.
Similar content being viewed by others
References
Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., and Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008.
Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., and Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712.
Chung C.T., Niemela S.L., and Miller R.H. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86, 2172–2175.
Datsenko K.A. and Wanner B.L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645.
Ellis H.M., Yu D., DiTizio T., and Court D.L. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98, 6742–6746.
Esvelt K.M. and Wang H.H. 2013. Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 9, 6–1.
Garneau J.E., Dupuis M.È., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., and Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71.
Gasiunas G., Barrangou R., Horvath P., and Siksnys V. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 109, E2579–86.
Gibson D.G., Benders G.A., Andrews-Pfannkoch C., Denisova E.A., Baden-Tillson H., Zaveri J., Stockwell T.B., Brownley A., Thomas D.W., Algire M.A., and et al. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220.
Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C., and et al. 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734.
Hu S., Fu J., Huang F., Ding X., Stewart A.F., Xia L., and Zhang Y. 2014. Genome engineering of Agrobacterium tumefaciens using the lambda Red recombination system. Appl. Microbiol. Biotechnol. 98, 2165–2172.
Isaacs F.J., Carr P.A., Wang H.H., Lajoie M.J., Sterling B., Kraal L., Tolonen A.C., Gianoulis T.A., Goodman D.B., Reppas N.B., and et al. 2011. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 333, 348–353.
Kulkarni S.K.S. and Stahl F.W.F. 1989. Interaction between the sbcC gene of Escherichia coli and the gam gene of phage lambda. Genetics 123, 249–253.
Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J., and et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148.
Moerschell R.P., Tsunasawa S., and Sherman F. 1988. Transformation of yeast with synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 85, 524–528.
Mullis K.B. 1991. The polymerase chain reaction in an anemic mode: how to avoid cold oligodeoxyribonuclear fusion. PCR Methods Appl. 1, 1–4.
Murphy K.C. 1991. Lambda Gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J. Bacteriol. 173, 5808–5821.
Murphy K.C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180, 2063–2071.
Muyrers J.P.J., Zhang Y.Y., Testa G.G., and Stewart A.F.A. 1999. Rapid modification of bacterial artificial chromosomes by ETrecombination. Nucleic Acids Res. 27, 1555–1557.
New England BioLabs Gibson Assembly® Protocol (E5510). https://www.neb.com/protocols/2012/12/11/gibson-assembly-protocol-e5510?device=pdf.
Orr-Weaver T.L., Szostak J.W., and Rothstein R.J. 1983. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 101, 228–245.
Rashtchian A. 1995. Novel methods for cloning and engineering genes using the polymerase chain reaction. Curr. Opin. Biotechnol. 6, 30–36.
Ryu Y.S., Biswas R.K., Shin K., Parisutham V., Kim S.M., and Lee S.K. 2014. A simple and effective method for construction of Escherichia coli strains proficient for genome engineering. PLoS ONE 9, e942–6.
Sambrook J. and Russell D.W. 2006. The Inoue method for preparation and transformation of competent E. coli: “ultra-competent” cells. CSH Protoc. 2006, 1–6.
Seidman C.E., Struhl K., Sheen J., and Jessen T. 2001. Introduction of plasmid DNA into cells. Current Protocols in Molecular Biology, John Wiley & Sons, Inc., Hoboken, NJ, USA.
TAKARA Bio. 2007. PrimeStarMax mutagenesis kit 1–3.
Thomason L.C., Sawitzke J.A., Li X., Costantino N., and Court D.L. 2014. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 106, 1.16.1–1.16.39.
Ting J.T. and Feng G. 2014. Recombineering strategies for developing next generation BAC transgenic tools for optogenetics and beyond. Front Behav. Neurosci. 8, 1–1.
Wang H.H., Isaacs F.J., Carr P.A., Sun Z.Z., Xu G., Forest C.R., and Church G.M. 2009. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898.
Yon J. and Fried M. 1989. Precise gene fusion by PCR. Nucleic Acids Res. 17, 48–5.
Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., and Court D.L. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97, 5978–5983.
Zhang Y.Y., Buchholz F.F., Muyrers J.P.J., and Stewart A.F.A. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20, 123–128.
Zhang Y., Werling U., and Edelmann W. 2012. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 40, e55–e55.
Zheng L. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115–e115.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nakayama, H., Shimamoto, N. Modern and simple construction of plasmid: Saving time and cost. J Microbiol. 52, 891–897 (2014). https://doi.org/10.1007/s12275-014-4501-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-014-4501-6