Abstract
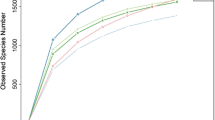
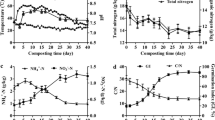
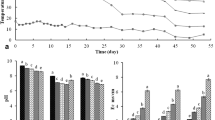
The concentration of major odor-causing compounds including phenols, indoles, short-chain fatty acids (SCFAs) and branched chain fatty acids (BCFAs) in response to the addition of powdered horse radish (PHR) and spent mushroom compost (SMC) was compared with control non-treated slurry (CNS) samples. A total of 97,465 rDNAs sequence reads were generated from three different samples (CNS, n = 2; PHR, n = 3; SMC, n = 3) using bar-coded pyrosequencing. The number of operational taxonomic units (OTUs) was lower in the PHR slurry compared with the other samples. A total of 11 phyla were observed in the slurry samples, while the phylogenetic analysis revealed that the slurry microbiome predominantly comprised members of the Bacteroidetes, Firmicutes, and Proteobacteria phyla. The rarefaction analysis showed the bacterial species richness varied among the treated samples. Overall, at the OTU level, 2,558 individual genera were classified, 276 genera were found among the three samples, and 1,832 additional genera were identified in the individual samples. A principal component analysis revealed the differences in microbial communities among the CNS, PHR, and SMC pig slurries. Correlation of the bacterial community structure with the Kyoto Encyclopedia of Genes and Genomes (KEGG) predicted pathways showed that the treatments altered the metabolic capabilities of the slurry microbiota. Overall, these results demonstrated that the PHR and S MC treatments significantly reduced the malodor compounds in pig slurry (P < 0.05).
Similar content being viewed by others
References
Ahn J.H., Hong I.P., Bok J.I., Kim B.Y., Song J., and Weon H.Y. 2012a. Pyrosequencing analysis of the bacterial communities in the guts of honey bees Apis cerana and Apis mellifera in Korea. J. Microbiol. 50, 735–745.
Ahn J.H., Kim B.Y., Song J., and Weon H.Y. 2012b. Effects of PCR cycle number and DNA polymerase type on the 16S rRNA gene pyrosequencing analysis of bacterial communities. J. Microbiol. 50, 1071–1074.
Allen H.K., Looft T., Bayles D.O., Humphrey S., Levine U.Y., Alt D., and Stanton T.B. 2011. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio. 2, e00260–11.
Ariesyady H.D., Ito T., and Okabe S. 2007. Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res. 41, 1554–1568.
Bacosa H., Suto K., and Inoue C. 2010. Preferential degradation of aromatic hydrocarbons in kerosene by a microbial consortium. Int. Biodeterior. Biodegrad. 64, 702–710.
Bacosa H.P., Suto K., and Inoue C. 2011. Preferential utilization of petroleum oil hydrocarbon components by microbial consortia reflects degradation pattern in aliphatic-aromatic hydrocarbon binary mixtures. World J. Microbiol. Biotechnol. 27, 1109–1117.
Blanes-Vidal V., Suh H., Nadimi E.S., Lofstrom P., Ellermann T., Andersen H.V., and Schwartz J. 2012. Residential exposure to outdoor air pollution from livestock operations and perceived annoyance among citizens. Environ. Int. 40, 44–50.
Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I., Knight R., Mills D.A., and Caporaso J.G. 2013. Qualityfiltering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 10, 57–59.
Bowers K.J., Mesbah N.M., and Wiegel J. 2009. Biodiversity of poly-extremophilic bacteria: Does combining the extremes of high salt, alkaline ph and elevated temperature approach a physico-chemical boundary for life? Saline Syst. 5, 9.
Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., and et al. 2010a. Qiime allows analysis of highthroughput community sequencing data. Nat. Methods 7, 335–336.
Caporaso J.G., Bittinger K., Bushman F.D., DeSantis T.Z., Andersen G.L., and Knight R. 2010b. Pynast: A flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267.
Chen S. and Dong X. 2005. Proteiniphilum acetatigenes gen Nov., sp. Nov., from a uasb reactor treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 55, 2257–2261.
Clarke K.R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143.
Cottrell M.T. and Kirchman D.L. 2000. Natural assemblages of marine proteobacteria and members of the cytophaga-flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66, 1692–1697.
Downes J., Vartoukian S.R., Dewhirst F.E., Izard J., Chen T., Yu W.H., Sutcliffe I.C., and Wade W.G. 2009. Pyramidobacter piscolens gen Nov., sp. Nov., a member of the phylum ‘synergistetes’ isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 59, 972–980.
Edgar R.C. 2010. Search and clustering orders of magnitude faster than blast. Bioinformatics 26, 2460–2461.
Edgar R.C., Haas B.J., Clemente J.C., Quince C., and Knight R. 2011. Uchime improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200.
Flint H.J., Scott K.P., Duncan S.H., Louis P., and Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306.
Fox L.R., Purves W.K., and Nakada H.I. 1965. The role of horseradish peroxidase in indole-3-acetic acid oxidation. Biochemistry 4, 2754–2763.
Govere E.M., Tonegawa M., Bruns M.A., Wheeler E.F., Heinemann P.H., Kephart K.B., and Dec J. 2005. Deodorization of swine manure using minced horseradish roots and peroxides. J. Agricult. Food Chem. 53, 4880–4889.
Govere E.M., Tonegawa M., Bruns M.A., Wheeler E.F., Kephart K.B., Voigt J.W., and Dec J. 2007. Using minced horseradish roots and peroxides for the deodorization of swine manure: A pilot scale study. Bioresour. Technol. 98, 1191–1198.
Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S.K., Sodergren E., and et al. 2011. Chimeric 16S rRNA sequence formation and detection in sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504.
Hatamoto M., Imachi H., Yashiro Y., Ohashi A., and Harada H. 2007. Diversity of anaerobic microorganisms involved in long-chain fatty acid degradation in methanogenic sludges as revealed by RNA-based stable isotope probing. Appl. Environ. Microbiol. 73, 4119–4127.
Hawe S.M., Walker N., and Moss B.W. 1992. The effects of dietary fiber, lactose and antibiotic on the levels of skatole and indole in feces and subcutaneous fat in growing pigs. Anim. Prod. 54, 413–419.
Hobbs P.J., Misselbrook T.H., and Cumby T.R. 1999. Production and emission of odours and gases from ageing pig waste. J. Agric. Engng. Res. 72, 291–298.
Jensen M.T., Cox R.P., and Jensen B.B. 1995. 3-methylindole (skatole) and indole production by mixed populations of pig fecal bacteria. Appl. Environ. Microbiol. 61, 3180–3184.
Juteau P., Bisaillon J.G., Lepine F., Ratheau V., Beaudet R., and Villemur R. 2003. Improving the biotreatment of hydrocarbons- contaminated soils by addition of activated sludge taken from the wastewater treatment facilities of an oil refinery. Biodegradation 14, 31–40.
Kindaichi T., Ito T., and Okabe S. 2004. Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl. Environ. Microbiol. 70, 1641–1650.
Klibanov A.M., Tu T.M., and Scott K.P. 1983. Peroxidase-catalyzed removal of phenols from coal-conversion waste waters. Science 221, 259–261.
Lamendella R., Domingo J.W., Ghosh S., Martinson J., and Oerther D.B. 2011. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 11, 1–3.
Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R., and et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821.
Le P.D., Aarnink A.J.A., Jongbloed A.W., Van der Peet-Schwering C.M.C., Ogink N.W.M., and Verstegen M.W.A. 2008. Interactive effects of dietary crude protein and fermentable carbohydrate levels on odour from pig manure. Livest. Sci. 114, 48–61.
Leng J., Xie L., Zhu R., Yang S., Gou X., Li S., and Mao H. 2011. Dominant bacterial communities in the rumen of gayals (bos frontalis), yaks (bos grunniens) and yunnan yellow cattle (bos taurs) revealed by denaturing gradient gel electrophoresis. Mol. Biol. Rep. 38, 4863–4872.
Leser T.D., Amenuvor J.Z., Jensen T.K., Lindecrona R.H., Boye M., and Moller K. 2002. Culture-independent analysis of gut bacteria: The pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68, 673–690.
Leung M.Y.K., Fung K.P., and Choy Y.M. 1997. The isolation and characterization of an immunomodulatory and anti-tumor polysaccharide preparation from Flammulina velutipes. Immunopharmacology 35, 255–263.
Li C.Y., Liu J.X., Wang Y.Z., Wu Y.M., Wang J.K., and Zhou Y.Y. 2009. Influence of differing carbohydrate sources on ltryptophan metabolism by porcine fecal microbiota studied in vitro. Livest. Sci. 120, 43–50.
Liao C.M. and Bundy D.S. 1994. Bacteria additives to the changes in gaseous mass-transfer from stored swine manure. J. Environ. Sci. Heal. B. 29, 1219–1249.
Looft T., Johnson T.A., Allen H.K., Bayles D.O., Alt D.P., Stedtfeld R.D., Sul W.J., Stedtfeld T.M., Chai B., Cole J.R., and et al. 2012. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 109, 1691–1696.
Lozupone C. and Knight R. 2005. Unifrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235.
Lozupone C., Lladser M.E., Knights D., Stombaugh J., and Knight R. 2011. Unifrac: An effective distance metric for microbial community comparison. ISME J. 5, 169–172.
Ma Y.F., Zhang Y., Zhang J.Y., Chen D.W., Zhu Y., Zheng H., Wang S.Y., Jiang C.Y., Zhao G.P., and Liu S.J. 2009. The complete genome of comamonas testosteroni reveals its genetic adaptations to changing environments. Appl. Environ. Microbiol. 75, 6812–6819.
Mackie R.I., Stroot P.G., and Varel V.H. 1998. Biochemical identification and biological origin of key odor components in livestock waste. J. Anim. Sci. 76, 1331–1342.
Mao S.Y., Zhang R.Y., Wang D.S., and Zhu W.Y. 2012. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet. Res. 8, 2–7.
Marchesi J.R. 2010. Prokaryotic and eukaryotic diversity of the human gut. Adv. Appl. Microbiol. 72, 43–62.
McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., and Hugenholtz P. 2012a. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618.
McDonald J.E., Houghton J.N., Rooks D.J., Allison H.E., and McCarthy A.J. 2012b. The microbial ecology of anaerobic cellulose degradation in municipal waste landfill sites: Evidence of a role for fibrobacters. Environ. Microbiol. 14, 1077–1087.
Middelbos I.S., Boler B.M.V., Qu A., White B.A., Swanson K.S., and Fahey G.C. 2010. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS ONE 5, e9768.
Miller D.N. and Varel V.H. 2003. Swine manure composition affects the biochemical origins, composition, and accumulation of odorous compounds. J. Anim. Sci. 81, 2131–2138.
Morawski B., Quan S., and Arnold F.H. 2001. Functional expression and stabilization of horseradish peroxidase by directed evolution in Saccharomyces cerevisiae. Biotechnol. Bioeng. 76, 99–107.
Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B., and et al. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79.
Neifar M., Jaouani A., Martínez M.J., and Penninckx M.J. 2012. Comparative study of olive oil mill wastewater treatment using free and immobilized Coriolopsis polyzona and Pycnoporus coccineus. J. Microbiol. 50, 746–753.
Ni J., Yan Q., and Yu Y. 2013. How much metagenomic sequencing is enough to achieve a given goal? Sci Rep. 3, 19–8.
Niu B., Fu L., Sun S., and Li W. 2010. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 11, 1–7.
Oh J., Kim B.K., Cho W.S., Hong S.G., and Kim K.M. 2012. Pyro-Trimmer: a software with GUI for pre-processing 454 amplicon sequences. J. Microbiol. 50, 766–769.
Overland M., Kjos N.K., Fauske A.K., Teige J., and Sorum H. 2011. Easily fermentable carbohydrates reduce skatole formation in the distal intestine of entire male pigs. Livest. Sci. 140, 206–217.
Parker D.B. 2008. Reduction of odor and voc emissions from a dairy lagoon. Appl. Eng. Agric. 24, 647–655.
Parker D.B., Cai L., Kim K.H., Hales K.E., Spiehs M.J., Woodbury B.L., Atkin A.L., Nickerson K.W., and Patefield K.D. 2012. Reducing odorous voc emissions from swine manure using soybean peroxidase and peroxides. Bioresour. Technol. 124, 95–104.
Roling W.F., Ferrer M., and Golyshin P.N. 2010. Systems approaches to microbial communities and their functioning. Curr. Opin. Biotechnol. 21, 532–538.
Samanta A.K., Torok V.A., Percy N.J., Abimosleh S.M., and Howarth G.S. 2012. Microbial fingerprinting detects unique bacterial communities in the faecal microbiota of rats with experimentally-induced colitis. J. Microbiol. 50, 218–225.
Savage D.C., Dubos R., and Schaedler R.W. 1968. The gastrointestinal epithelium and its autochthonous bacterial flora. J. Exp. Med. 127, 67–76.
Snaidr J., Amann R., Huber I., Ludwig W., and Schleifer K.H. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63, 2884–2896.
Snell-Castro R., Godon J.J., Delgenes J.P., and Dabert P. 2005. Characterisation of the microbial diversity in a pig manure storage pit using small subunit rdna sequence analysis. FEMS Microbiol. Ecol. 52, 229–242.
Sousa D.Z., Pereira M.A., Smidt H., Stams A.J., and Alves M.M. 2007. Molecular assessment of complex microbial communities degrading long chain fatty acids in methanogenic bioreactors. FEMS Microbiol. Ecol. 60, 252–265.
Subair S., Fyles J., and O’Halloran I. 1999. Ammonia volatilization from liquid hog manure amended with paper products in the laboratory. J. Environ. Qual. 28, 202–207.
Sutton A.L., Kephart K.B., Verstegen M.W., Canh T.T., and Hobbs P.J. 1999. Potential for reduction of odorous compounds in swine manure through diet modification. J. Anim. Sci. 77, 430–439.
Tajima K., Aminov R.I., Nagamine T., Ogata K., Nakamura M., Matsui H., and Benno Y. 1999. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol. Ecol. 29, 159–169.
Tonegawa M., Dec J., and Bollag J.M. 2003. Use of additives to enhance the removal of phenols from water treated with horseradish and hydrogen peroxide. J. Environ. Qual. 32, 1222–1227.
Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., and Gordon J.I. 2007. The human microbiome project. Nature 449, 804–810.
Wang Q., Garrity G.M., Tiedje J.M., and Cole J.R. 2007a. Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267.
Wang H.F., Zhu W.Y., Yao W., and Liu J.X. 2007b. Dgge and 16S rDNA sequencing analysis of bacterial communities in colon content and feces of pigs fed whole crop rice. Anaerobe 13, 127–133.
Williams A.G. and Evans M.R. 1981. Storage of piggery slurry. Agr. Wastes 3, 311–321.
Williams B., McMullan J., and McCahey S. 2001. An initial assessment of spent mushroom compost as a potential energy feedstock. Bioresour. Technol. 79, 227–230.
Ye F.X., Zhu R.F., and Li Y. 2009. Deodorization of swine manure slurry using horseradish peroxidase and peroxides. J. Hazard. Mater. 167, 148–153.
Ziemer C.J., Kerr B.J., Trabue S.L., Stein H., Stahl D.A., and Davidson S.K. 2009. Dietary protein and cellulose effects on chemical and microbial characteristics of swine feces and stored manure. J. Environ. Qual. 38, 2138–2146.
Author information
Authors and Affiliations
Corresponding authors
Additional information
These authors contributed equally to this work
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hwang, OH., Raveendar, S., Kim, YJ. et al. Deodorization of pig slurry and characterization of bacterial diversity using 16S rDNA sequence analysis. J Microbiol. 52, 918–929 (2014). https://doi.org/10.1007/s12275-014-4251-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-014-4251-5