Abstract
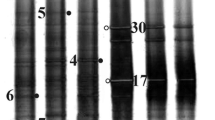
The microbial diversity and biogeochemical potential associated with a northern Saskatchewan uranium mine water-tailings interface was examined using culture-dependent and -independent techniques. Morphologically-distinct colonies from uranium mine water-tailings and a reference lake (MC) obtained using selective and non-selective media were selected for 16S rRNA gene sequencing and identification, revealing that culturable organisms from the uranium tailings interface were dominated by Firmicutes and Betaproteobacteria; whereas, MC organisms mainly consisted of Bacteroidetes and Gammaproteobacteria. Ion Torrent (IT) 16S rRNA metagenomic analysis carried out on extracted DNA from tailings and MC interfaces demonstrated the dominance of Firmicutes in both of the systems. Overall, the tailings-water interface environment harbored a distinct bacterial community relative to the MC, reflective of the ambient conditions (i.e., total dissolved solids, pH, salinity, conductivity, heavy metals) dominating the uranium tailings system. Significant correlations among the physicochemical data and the major bacterial groups present in the tailings and MC were also observed. Presence of sulfate reducing bacteria demonstrated by culture-dependent analyses and the dominance of Desulfosporosinus spp. indicated by Ion Torrent analyses within the tailings-water interface suggests the existence of anaerobic microenvironments along with the potential for reductive metabolic processes.
Similar content being viewed by others
References
Akob, D.M., Mills, H.J., and Kostka, J.E. 2007. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59, 95–107.
Benedetto, J.S., de Almeida, S.K., Gomes, H.A., Vazoller, R.F., and Ladeira, A.C.Q. 2005. Monitoring of sulfate-reducing bacteria in acid water from uranium mines. Miner. Eng. 18, 1341–1343.
Bondici, V.F., Lawrence, J.R., Khan, N.H., Hill, J.E., Yergeau, E., Wolfaardt, G.M., Warner, J., and Korber, D.R. 2013. Microbial communities in low permeability, high pH uranium mine tailings: characterization and potential effects. J. Appl. Microbiol. 114, 1671–1686.
Bowers, K.J., Mesbah, N.M., and Wiegel, J. 2009. Biodiversity of poly-extremophilic Bacteria: Does combining the extremes of high salt, alkaline pH and elevated temperature approach a physic-chemical boundary for life? Saline Syst. 5, 9.
Burkhardt, E.M., Akob, D.M., Bischoff, S., Sitte, S., Kostka, J.O., Banerjee, D., Scheinost, A.C., and Kusel, K. 2010. Impact of biostimulated redox processes on metal dynamics in an iron-rich creek soil of a former uranium mining area. Environ. Sci. Technol. 44, 177–183.
Dib, J.R., Annika, W., Neumann, A., Ordoñez, O., Estévez, M.C., and Farías, M.E. 2009. Isolation of bacteria from remote high altitude Andean Lakes able to grow in the presence of antibiotics. Recent Patents Anti-Infective Drug Discov. 4, 66–76.
Duckworth, A.W., Grant, W.D., Jones, B.E., and van Steenbergen, R. 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol. Ecol. 19, 181–191.
Ellis, R.J.M., Morgan, P., Weightman, A.J., and Fry, J.C. 2003. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl. Environ. Microbiol. 69, 3223–3230.
Herlihy, A.T. and Mills, A.L. 1985. Sulfate reduction in freshwater sediments receiving acid mine drainage. Appl. Environ. Microbiol. 49, 179–186.
Hirkala, D.L.M. and Germida, J.J. 2004. Field and soil microcosm studies on the survival and conjugation of a Pseudomonas putida strain bearing a recombination plasmid, pADPTel. Can. J. Microbiol. 50, 595–604.
Islam, E., Dhal, P.K., Kazy, S.K., and Sar, P. 2011. Molecular analysis of bacterial communities in uranium ores and surrounding soils from Banduhurang open cast uranium mine, India: A comparative study. J. Environ. Sci. Heal. A. 46, 271–280.
Islam, E. and Sar, P. 2011. Culture-dependent and -independent molecular analysis of the bacterial community within uranium ore. J. Basic Microbiol. 51, 372–384.
Jain, D.K. 1995. Evaluation of semisolid Postgate’s B medium for enumerating sulfate-reducing bacteria. J. Microbiol. Methods 22, 27–38.
Jones, B.E., Grant, W.D., Duckworth, A.W., and Owenson, G.G. 1998. Microbial diversity of soda lakes. Extremophiles 2, 191–200.
Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120.
Landa, E.R., Phillips, E.J.P., and Lovley, D.R. 1991. Release of (super 226) Ra from uranium mill tailings by microbial Fe(III) reduction. Appl. Geochem. 6, 647–652.
Ley, R.E., Turnbaugh, P.J., Klein, S., and Gordon, J.T. 2006. Microbial ecology: Human gut microbes associated with obesity. Nature 444, 1022–1023.
Lovley, D.R. and Phillips, E.J.P. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58, 850–856.
Lovley, D.R., Widman, P.K., Woodward, J.C., and Phillips, E.J. 1993. Reduction of uranium by cytochrome C3 of Desulfovibrio vulgaris. Appl. Environ. Microbiol. 59, 3572–3576.
Ma, Y., Xue, Y., Grant, W.D., Collins, N.C., Duckworth, A.W., van Steenbergen, R.P., and Jones, B.E. 2004. Alkalimonas amylolytica gen. nov., sp. nov., and Alkalimonas delamerensis gen. nov., sp. nov., novel alkaliphilic bacteria from soda lakes in China and East Africa. Extremophiles 8, 193–200.
Miller, C.L., Landa, E.R., and Updegraff, D.M. 1987. Ecological aspects of microorganisms inhabiting uranium mill tailings. Microb. Ecol. 14, 141–155.
Mills, A.L., Bell, P.E., and Herlihy, A.T. 1989. Microbes, sediments, and acidified waters: the importance of biological buffering, pp. 1–19. In Rao, S.S. (ed.), Microbial interactions in acid stressed aquatic ecosystems. CRC Press, Inc., Boca Raton, FL, USA.
Mitsui, H., Gorlach, K., Lee, H., Hattori, J.R., and Hattori, T. 1997. Incubation time and media requirements of culturable bacteria from different phylogenetic groups. J. Microbiol. Methods 30, 103–110.
Moldovan, B.J. and Hendry, M.J. 2005. Characterizing and quantifying controls on arsenic solubility over a pH range 1–11 in a uranium mill experiment. Environ. Sci. Technol. 39, 4913–4920.
Moreels, D., Garry, C., Garafola, C., Monteleone, D., Taghavi, S., Fitts, J.P., and van der Lelie, D. 2008. Microbial community dynamics in uranium contaminated subsurface sediments under biostimulated conditions with high nitrate and nickel pressure. Environ. Sci. Pollut. Res. 15, 481–491.
Natural Resources Canada. 2006. Canada’s uranium production and nuclear power. Nuclear Issue Briefing, Paper No. 3. Ottawa, Ontario, Canada.
Pyle, G.G., Swanson, S.M., and Lehmkuhl, D.M. 2002. Toxicity of uranium mine receiving waters to early life stage fathead minnows (Pimephales promelas) in the laboratory. Environ. Pollut. 116, 243–255.
Radeva, G. and Selenska-Pobell, S. 2005. Bacterial diversity in water samples from uranium wastes as demonstrated by 16S rDNA and ribosomal intergenic spacer amplification retrievals. Can. J. Microbiol. 51, 910–923.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Saskatchewan Mining Association. 2010. Saskatchewan Mining Association Fact Sheet. 2010. SMA, Regina, SK, Canada.
Selenska-Pobell, S. 2002. Diversity and activity of bacteria in uranium waste piles, pp. 225–254. In Keith-Roach, M.J. and Livens, F.R. (eds.), Interactions of microorganisms with radionuclides, Elsevier Sciences Ltd., Oxford, UK.
Selenska-Pobell, S., Kampf, G., Hemming, K., Radeva, G., and Satchanska, G. 2001. Bacterial diversity in soil samples from two uranium waste piles as determined by rep-APD, RISA and 16S rDNA retrieval. Antonie van Leeuwenheok 79, 149–161.
Shaw, S.A., Hendry, M.J., Esselfie-Dughan, J., Kotzer, T., and Wallschlager, D. 2011 Distribution, characterization, and geochemical controls of elements of concern in uranium mine tailings, Key Lake, Saskatchewan, Canada. Appl. Geochem. 26, 2044–2056.
Shelobolina, E.S., O’Neill, K., Finneran, K.T., Hayes, L.A., and Lovley, D.R. 2003. Potential for in situ bioremediation of a low-pH, high-nitrate uranium-contaminated groundwater. Soil Sediment Contam. 12, 865–884.
Singleton, R. Jr. 1993. The sulfate-reducing bacteria: an overview, pp. 1–20. In Odom, J.M. and Singleton, R. (eds.), The sulfate-reducing bacteria: contemporary perspectives. Springer Verlag, Inc., New York, N.Y., USA.
Sitte, J., Akob, M.D.M., Kaufmann, C., Finster, K., Banerjee, D., Burkhardt, E.M., Kostka, J.E., Scheinost, A.C., Büchel, G., and Küsel, K. 2010. Microbial links between sulfate reduction and metal retention in uranium- and heavy metal-contaminated soil. Appl. Environ. Microbiol. 76, 3143–3152.
Sorokin, D.Y., Banciu, H., van Loosdrecht, M., and Kuenen, J.G. 2003. Growth physiology and competitive interaction of obligately chemolithoautotrophic, haloalkaliphilic, sulphur-oxidizing bacteria from soda lakes. Extremophiles 7, 195–203.
Stackebrandt, E., Schumann, P., Schüler, E., and Hippe, H. 2003. Reclassification of Desulfotomaculum auripigmentum as Desulfosporosinus auripigmenti corrig., comb. nov. Int. J. Syst. Evol. Microbiol. 53, 1439–1443.
Suzuki, Y., Kelly, S.D., Kemner, K.M., and Banfield, J.F. 2002. Nanometre-sized products of uranium bioreduction. Nature 419, 134.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739.
Vatsurina, A., Badrutdinova, D., Schumann, P., and Spring, S.M. 2008. Desulfosporosinus hippei sp. nov., a mesophilic sulfate-reducing bacterium isolated from permafrost. Int. J. Syst. Evol. Microbiol. 58, 1228–1232.
Wagner, D. 2008. Microbial communities and processes in arctic permafrost environments, pp. 133–154. In Dion, P. and Chandra, S.N. (eds.), Microbiology of extreme soils. Soil Biology. vol. 13. Springer, Berlin, Germany.
Wolfaardt, G.M., Hendry, M.J., and Korber, D.R. 2008. Microbiology of saturated, high pH uranium-mine tailings, Saskatchewan, Canada. Can. J. Microbiol. 54, 932–940.
Yergeau, E., Lawrence, J.R., Sanschagrin, S., Waiser, M.J., Korber, D.R., and Greer, C.W. 2012. Next-generation sequencing of microbial communities in the Athabasca River and its tributaries in relation to oil sands mining activities. Appl. Environ. Microbiol. 78, 7626–7637.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khan, N.H., Bondici, V.F., Medihala, P.G. et al. Bacterial diversity and composition of an alkaline uranium mine tailings-water interface. J Microbiol. 51, 558–569 (2013). https://doi.org/10.1007/s12275-013-3075-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-013-3075-z