Abstract
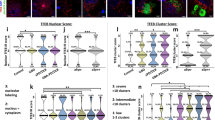
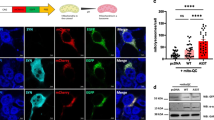
Huntington’s disease (HD) is a deadly neurodegenerative disease with abnormal expansion of CAG repeats in the huntingtin gene. Mutant Huntingtin protein (mHTT) forms abnormal aggregates and intranuclear inclusions in specific neurons, resulting in cell death. Here, we tested the ability of a natural heat-shock protein 90 inhibitor, Gedunin, to degrade transfected mHTT in Neuro-2a cells and endogenous mHTT aggregates and intranuclear inclusions in both fibroblasts from HD patients and neurons derived from induced pluripotent stem cells from patients. Our data showed that Gedunin treatment degraded transfected mHTT in Neuro-2a cells, endogenous mHTT aggregates and intranuclear inclusions in fibroblasts from HD patients, and in neurons derived from induced pluripotent stem cells from patients in a dose- and time-dependent manner, and its activity depended on the proteasomal pathway rather than the autophagy route. These findings also showed that although Gedunin degraded abnormal mHTT aggregates and intranuclear inclusions in cells from HD patient, it did not affect normal cells, thus providing a new perspective for using Gedunin to treat HD.






Similar content being viewed by others
References
MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72: 971–983.
Kieburtz K, Reilmann R, Olanow CW. Huntington’s disease: current and future therapeutic prospects. Mov Disord 2018, 33: 1033–1041.
Squitieri F, Griguoli A, Capelli G, Porcellini A, D’Alessio B. Epidemiology of Huntington disease: first post-HTT gene analysis of prevalence in Italy. Clin Genet 2016, 89: 367–370.
Louis ED, Lee P, Quinn L, Marder K. Dystonia in Huntington’s disease: prevalence and clinical characteristics. Mov Disord 1999, 14: 95–101.
Walker FO. Huntington’s disease. Semin Neurol 2007, 27: 143–150.
Slaughter JR, Martens MP, Slaughter KA. Depression and Huntington’s disease: prevalence, clinical manifestations, etiology, and treatment. CNS Spectr 2001, 6: 306–326.
Orth M, Handley OJ, Schwenke C, Dunnett SB, Craufurd D, Ho AK, et al. Observing Huntington’s disease: the European Huntington’s disease network’s REGISTRY. PLoS Curr 2010, 2: RRN1184.
Aylward EH, Nopoulos PC, Ross CA, Langbehn DR, Pierson RK, Mills JA, et al. Longitudinal change in regional brain volumes in prodromal Huntington disease. J Neurol Neurosurg Psychiatry 2011, 82: 405–410.
Saudou F, Humbert S. The biology of Huntingtin. Neuron 2016, 89: 910–926.
Li HL, Zhang YB, Wu ZY. Development of research on Huntington disease in China. Neurosci Bull 2017, 33: 312–316.
DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, et al. Aggregation of Huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 1997, 277: 1990–1993.
Lee JM, Ramos EM, Lee JH, Gillis T, Mysore JS, Hayden MR, et al. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology 2012, 78: 690–695.
Zhang WY, Gu ZL, Liang ZQ, Qin ZH. Mitochondrial dysfunction and Huntington disease. Neurosci Bull 2006, 22: 129–136.
Fisher ER, Hayden MR. Multisource ascertainment of Huntington disease in Canada: prevalence and population at risk. Mov Disord 2014, 29: 105–114.
Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol 2010, 11: 515–528.
Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer 2005, 5: 761–772.
Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K. A Screen for enhancers of clearance identifies Huntingtin as a heat shock protein 90 (Hsp90) client protein. J Biol Chem 2012, 287: 1406–1414.
Luo WJ, Sun WL, Taldone T, Rodina A, Chiosis G. Heat shock protein 90 in neurodegenerative diseases. Mol Neurodegener 2010, 5: 24.
He WT, Xue W, Gao YG, Hong JY, Yue HW, Jiang LL, et al. HSP90 recognizes the N-terminus of Huntingtin involved in regulation of Huntingtin aggregation by USP19. Sci Rep 2017, 7: 14797.
Tokui K, Adachi H, Waza M, Katsuno M, Minamiyama M, Doi H, et al. 17-DMAG ameliorates polyglutamine-mediated motor neuron degeneration through well-preserved proteasome function in an SBMA model mouse. Hum Mol Genet 2009, 18: 898–910.
Patwardhan CA, Fauq A, Peterson LB, Miller C, Blagg BS, Chadli A. Gedunin inactivates the co-chaperone p23 protein causing cancer cell death by apoptosis. J Biol Chem 2013, 288: 7313–7325.
Ma L, Hu B, Liu Y, Vermilyea Scott C, Liu H, Gao L, et al. Human embryonic stem cell-derived gaba neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell 2012, 10: 455–464.
van Hagen M, Piebes DGE, de Leeuw WC, Vuist IM, van Roon-Mom WMC, Moerland PD, et al. The dynamics of early-state transcriptional changes and aggregate formation in a Huntington’s disease cell model. BMC Genomics 2017, 18: 373.
Legleiter J, Lotz GP, Miller J, Ko J, Ng C, Williams GL, et al. Monoclonal antibodies recognize distinct conformational epitopes formed by polyglutamine in a mutant Huntingtin fragment. J Biol Chem 2009, 284: 21647–21658.
Weiss A, Grueninger S, Abramowski D, Giorgio FP, Lopatin MM, Rosas HD, et al. Microtiter plate quantification of mutant and wild-type Huntingtin normalized to cell count. Anal Biochem 2011, 410: 304–306.
Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006, 443: 780–786.
Button RW, Luo SQ, Rubinsztein DC. Autophagic activity in neuronal cell death. Neurosci Bull 2015, 31: 382–394.
Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 1998, 8: 397–403.
Shacka JJ, Klocke BJ, Roth KA. Autophagy, bafilomycin and cell death: the “a-B-cs” of plecomacrolide-induced neuroprotection. Autophagy 2006, 2: 228–230.
Brooks E, Arrasate M, Cheung K, Finkbeiner SM. Using antibodies to analyze polyglutamine stretches. Methods Mol Biol 2004, 277: 103–128.
Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, et al. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci 1999, 19: 2522–2534.
Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, et al. Polyglutamine-expanded Huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 2017, 94: 48–57e44.
Rodriguez-Lebron E, Paulson HL. Allele-specific RNA interference for neurological disease. Gene Ther 2006, 13: 576–581.
Aguiar S, van der Gaag B, Cortese FAB. RNAi mechanisms in Huntington’s disease therapy: siRNA versus shRNA. Transl Neurodegener 2017, 6: 30.
Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 2018, 24: 939–946.
Yang S, Chang R, Yang H, Zhao T, Hong Y, Kong HE, et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J Clin Invest 2017, 127: 2719–2724.
Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 2018, 24: 927.
Becher MW, Kotzuk JA, Sharp AH, Davies SW, Bates GP, Price DL, et al. Intranuclear neuronal inclusions in Huntington’s disease and dentatorubral and pallidoluysian atrophy: correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol Dis 1998, 4: 387–397.
Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science 1995, 269: 407–410.
Flierl A, Oliveira LM, Falomir-Lockhart LJ, Mak SK, Hesley J, Soldner F, et al. Higher vulnerability and stress sensitivity of neuronal precursor cells carrying an alpha-synuclein gene triplication. PLoS One 2014, 9: e112413.
Acknowledgements
We thank all members of the HD families for trusting us and supporting us to the end of this work. We also thank Dr. Su-Chun Zhang for helping with this project. This work was supported by the National Key Research and Development Program of China (2018YFA0108004) and the National Natural Science Foundation of China (81271259).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors claim that there are no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, W., Xie, J., Qiang, Q. et al. Gedunin Degrades Aggregates of Mutant Huntingtin Protein and Intranuclear Inclusions via the Proteasomal Pathway in Neurons and Fibroblasts from Patients with Huntington’s Disease. Neurosci. Bull. 35, 1024–1034 (2019). https://doi.org/10.1007/s12264-019-00421-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12264-019-00421-5