Abstract
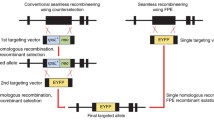
Recombineering has been developed to modify bacterial artificial chromosome (BAC) via homologous recombination. Nevertheless, as a screening strategy to identify the correct clone was not properly developed, it was difficult to obtain a correct clone within a limited time period. To address these issues, we developed a new screening method (a gain & loss screening system) that enables the efficient identification of the recombineered clone. Simple inoculation of cells into LB medium with appropriate antibiotics visually revealed the positive clones within 24 h. DNA sequencing confirmed 100% accuracy of this screening method by showing that all positive clones exhibited recombinant sequences. Furthermore, our new method allowed us to complete the entire procedure consisting of 1st recombineering, flip-out and 2nd recombineering in just 13 days. Overall, our new strategy may provide a new avenue for BAC recombineering, as evidenced by markedly increased accuracy and subsequently shortened recombineering duration.
Similar content being viewed by others
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- GOI:
-
gene-of-interest
- HR:
-
homology region
- KanR :
-
kanamycin resistant gene
- CamR :
-
chloramphenicol resistant gene
- AmpR :
-
ampicillin resistant gene
- NeoR :
-
neomycin resistance gene
- Kan:
-
kanamycin
- Cam:
-
chloramphenicol
- Amp:
-
ampicillin
- BTV:
-
BAC targeting vector
- BTC:
-
BAC targeting cassette
- CTV:
-
Cam targeting vector
- CTC:
-
Cam targeting cassette
- Fwd:
-
forward
- Rev:
-
reverse
- FLP :
-
flippase
- SOC:
-
Super Optimal broth with Catabolite repression
- DW:
-
Distilled Water
- OD:
-
optimal density
References
Wurm, F. M. (2004) Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 22: 1393–1398.
Rita Costa, A., M. Elisa Rodrigues, M. Henriques, J. Azeredo, and R. Oliveira (2010) Guidelines to cell engineering for monoclonal antibody production. Eur. J. Pharm. Biopharm. 74: 127–138.
Kunert, R. and E. Casanova (2013) Recent advances in recombinant protein production: BAC-based expression vectors, the bigger the better. Bioengineered. 4: 258–261.
Giraldo, P. and L. Montoliu (2001) Size matters: Use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 10: 83–103.
Sparwasser, T. and G. Eberl (2007) BAC to immunology—bacterial artificial chromosome-mediated transgenesis for targeting of immune cells. Immunology. 121: 308–313.
Van Keuren, M. L., G. B. Gavrilina, W. E. Filipiak, M. G. Zeidler, and T. L. Saunders (2009) Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic Res. 18: 769–785.
Deal, K. K., V. A. Cantrell, R. L. Chandler, T. L. Saunders, D. P. Mortlock, and E. M. Southard-Smith (2006) Distant regulatory elements in a Sox10-beta GEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev. Dyn. 235: 1413–1432.
Probst, F. J., R. A. Fridell, Y. Raphael, T. L. Saunders, A. Wang, Y. Liang, R. J. Morell, J. W. Touchman, R. H. Lyons, K. Noben-Trauth, T. B. Friedman, and S. A. Camper (1998) Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 280: 1444–1447.
Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart (1998) A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20: 123–128.
Muyrers, J. P., Y. Zhang, V. Benes, G. Testa, W. Ansorge, and A. F. Stewart (2000) Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 1: 239–243.
Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland (2001) A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 73: 56–65.
Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 97: 5978–5983.
Zhang, Y., C. Riesterer, A. M. Ayrall, F. Sablitzky, T. D. Littlewood, and M. Reth (1996) Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 24: 543–548.
Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland (2005) Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36.
Suster, M. L., G. Abe, A. Schouw, and K. Kawakami (2011) Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6: 1998–2021.
Balasubramanian, S., Y. Rajendra, L. Baldi, D. L. Hacker, and F. M. Wurm (2016) Comparison of three transposons for the generation of highly productive recombinant CHO cell pools and cell lines. Biotechnol. Bioeng. 113: 1234–1243.
Sharan, S. K., L. C. Thomason, S. G. Kuznetsov, and D. L. Court (2009) Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4: 206–223.
Kung, S. H., A. C. Retchless, J. Y. Kwan, and R. P. P. Almeida (2013) Effects of DNA size on transformation and recombination efficiencies in Xylella fastidiosa. Appl. Environ. Microbiol. 79: 1712–1717.
Jacobus, A. P. and J. Gross (2015) Optimal cloning of PCR fragments by homologous recombination in Escherichia coli. PLoS One. 10: e0119221.
Lee, P. Y., J. Costumbrado, C. Y. Hsu, and Y. H. Kim (2012) Agarose gel electrophoresis for the separation of DNA fragments. J. Vis. Exp. e3923.
Jahn, M., C. Vorpahl, T. Hübschmann, H. Harms, and S. Müller (2016) Copy number variability of expression plasmids determined by cell sorting and Droplet Digital PCR. Microb. Cell Fact. 15: 211.
Carreira-Rosario, A., S. Scoggin, N. A. Shalaby, N. D. Williams, P. R. Hiesinger, and M. Buszczak (2013) Recombineering homologous recombination constructs in Drosophila. J. Vis. Exp. e50346.
Liu, P., N. A. Jenkins, and N. G. Copeland (2003) A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13: 476–484.
Thomason, L. C., J. A. Sawitzke, X. Li, N. Costantino, and D. L. Court (2014) Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 106: 1.16.1–1.16.39.
Degryse, E. (1996) In vivo intermolecular recombination in Escherichia coli: application to plasmid constructions. Gene. 170: 45–50.
Dickinson, D. J., A. M. Pani, J. K. Heppert, C. D. Higgins, and B. Goldstein (2015) Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 200: 1035–1049.
Snounou, G. and A. D. Malcolm (1984) Supercoiling and the mechanism of restriction endonucleases. Eur. J. Biochem. 138: 275–280.
Asami, J., Y. U. Inoue, Y. W. Terakawa, S. F. Egusa, and T. Inoue (2011) Bacterial artificial chromosomes as analytical basis for gene transcriptional machineries. Transgenic Res. 20: 913–924.
Trivedi, R. N., P. Akhtar, J. Meade, P. Bartlow, M. M. Ataai, S. A. Khan, and M. M. Domach (2014) High-level production of plasmid DNA by Escherichia coli DH5α ΩsacB by introducing inc mutations. Appl. Environ. Microbiol. 80: 7154–7160.
Acknowledgements
This research was supported by priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A0304195411). This research was also supported by Research Assistance Program (2019) in the Incheon National University.
Author information
Authors and Affiliations
Contributions
MUK, KHW, SO and JTP conceived of and designed the experiments. MUK, YHL, JWK and SYH performed the experiments. MUK, KHW, SO and JTP wrote and edited the paper. All authors have read and approved the manuscript
Corresponding authors
Ethics declarations
Conflict of Interest The authors declare no competing financial interests.
Ethical Statement Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kuk, M.U., Lee, Y.H., Kim, J.W. et al. Rapid and Efficient BAC Recombineering: Gain & Loss Screening System. Biotechnol Bioproc E 26, 1023–1033 (2021). https://doi.org/10.1007/s12257-020-0382-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0382-1