Abstract
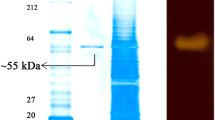
The alkaline α-amylase produced by Texcoconibacillus texcoconensis 13CCT strain was identified by random mutagenesis and confirmed by directed mutagenesis. A transposon mutagenesis approach was taken to identify the gene responsible for the degradation of starch in T. texcoconensis 13CCT strain. The deduced amino acids of the amy gene had a 99 % similarity with those of Bacillus selenitireducens MLS10 and 97 % with those of Paenibacillus curdlanolyticus YK9. The enzyme showed a maximum activity of 131.1 U/mL at 37 °C and pH 9.5 to 10.5. In situ activity of the enzyme determined by polyacrylamide gel electrophoresis showed only one band with amylolytic activity. This is the first report of a bacterium isolated from the extreme alkaline–saline soil of the former Lake Texcoco (Mexico) with amylolytic activity in alkaline conditions while its potential as a source of amylases for the industry is discussed.



Similar content being viewed by others
References
Alazard D, Badillo C, Fardeau ML, Cayol JL, Thomas P, Roldan T, Tholozan JL, Ollivier B (2007) Tindallia texcoconensis sp. nov., a new haloalkaliphilic bacterium isolated from lake Texcoco, Mexico. Extremophiles 11:33–39. doi:10.1007/s00792-006-0006-5
Annamalai N, Thavasi R, Vijayalakshmi S, Balasubramanian T (2011) Extraction, purification and characterization of thermostable, alkaline tolerant α-amylase from Bacillus cereus. Indian J Microbiol 51:424–429. doi:10.1007/s12088-011-0160-z
Bano S, Qader SA, Aman A, Syed MN, Azhar A (2011) Purification and characterization of novel α-amylase from Bacillus subtilis KIBGE HAS. AAPS PharmSciTech 12:255–261. doi:10.1208/s12249-011-9586-1
Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinforma 10:1–4. doi:10.1186/1471-2105-4-29
Cude WN, Mooney J, Tavanaei AA, Hadden MK, Frank AM, Gulvik CA, May AL, Buchan A (2012) Production of the antimicrobial secondary metabolite indigoidine contributes to competitive surface colonization by the marine roseobacter Phaeobacter sp. strain Y4I. Appl Environ Microbiol 78:4771–4780. doi:10.1128/AEM.00297-12
Demirkan E (2011) Production, purification, and characterization of α-amylase by Bacillus subtilis and its mutant derivates. Turk J Biol 35:705–712. doi:10.3906/biy-1009-113
Hagihara H, Igarashi K, Hayashi Y, Endo K, Ikawa-Kitayama K, Ozaki K, Kawai S, Ito S (2001) Novel α-amylase that is highly resistant to chelating reagents and chemical oxidants from the alkaliphilic Bacillus isolate KSM-K38. Appl Environ Microbiol 67:1744–1750. doi:10.1128/AEM.67.4.1744-1750.2001
Haki GD, Rakshit SK (2003) Developments in industrially important thermostable enzymes: a review. Bioresour Technol 89:17–34. doi:10.1016/S0960-8524(03)00033-6
Hernández-Montañez Z, Juárez-Montiel M, Velázquez-Ávila M, Cristiani-Urbina E, Hernández-Rodríguez C, Villa-Tanaca L, Chávez-Camarillo G (2012) Production and characterization of extracellular α-amylase produced by Wickerhamia sp. X-Fep. Appl Biochem Biotechnol 167:2117–2129. doi:10.1007/s12010-012-9736-2
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Larsen RA, Wilson MM, Guss AM, Metcalf WW (2002) Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi:10.1007/s00203-002-0442-2
Lin LL, Chyau CC, Hsu WH (1998) Production and properties of a raw-starch-degrading amylase from the thermophilic and alkaliphilic Bacillus sp. TS-23. Biotechnol Appl Biochem 28:61–68
Mahdavi A, Sajedi RH, Asghari SM, Taghdir M, Rassa M (2011) An analysis of temperature adaptation in cold active, mesophilic and thermophilic Bacillus α-amylases. Int J Biol Macromol 49:1038–1045. doi:10.1016/j.ijbiomac.2011.08.030
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Murakoshi Y, Makita T, Kato M, Kobayashi T (2012) Comparison and characterization of α-amylase inducers in Aspergillus nidulans based on nuclear localization of AmyR. Appl Microbiol Biotechnol 94:1629–1635. doi:10.1007/s00253-012-3874-x
Pascon RC, Bergamo RF, Spinelli RX, de Souza ED, Assis DM, Juliano L, Vallim MA (2011) Amylolytic microorganism from São Paulo zoo composting: isolation, identification, and amylase production. Enzyme Res 2011:679624. doi:10.4061/2011/679624
Richards GR, Vanderpool CK (2012) Induction of the pho regulon suppresses the growth defect of an Escherichia coli sgrS mutant, connecting phosphate metabolism to the glucose-phosphate stress response. J Bacteriol 194:2520–2530. doi:10.1128/JB.00009-12
Ruiz-Romero E, Coutiño-Coutiño MA, Valenzuela-Encinas C, López-Ramírez MP, Marsch R, Dendooven L (2013) Texcoconibacillus texcoconensis gen. nov. sp. nov., alkalophilic and halotolerant bacteria isolated from soil of the former lake Texcoco (Mexico). Int J Syst Evol Microbiol 63:3336–3341
Sammer UF, Reiher K, Spiteller D, Wensing A, Völksch B (2012) Assessment of the relevance of the antibiotic 2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine from Pantoea agglomerans biological control strains against bacterial plant pathogens. Microbiologyopen 1:438–449. doi:10.1002/mbo3.43
Schneider K, Asao M, Carter MS, Alber BE (2012) Rhodobacter sphaeroides uses a reductive route via propionyl coenzyme a to assimilate 3-hydroxypropionate. J Bacteriol 194:225–232. doi:10.1128/JB.05959-11
Shi W, Takano T, Liu S (2012) Isolation and characterization of novel bacterial taxa from extreme alkali-saline soil. World J Microbiol Biotechnol 28:2147–2157. doi:10.1007/s11274-012-1020-7
Sivaramakrishnan S, Gangadharan D, Nampoothiri KM (2006) α-Amylases from microbial sources—an over-view on recent developments. Food Technol Biotechnol 44:173–184
Tudor JJ, Davis JJ, Panichella M, Zwolak A (2008) Isolation of predation-deficient mutants of Bdellovibrio bacteriovorus by using transposon mutagenesis. Appl Environ Microbiol 74:5436–5443. doi:10.1128/AEM.00256-08
van den Burg B (2003) Extremophiles as a source for novel enzymes. Curr Opin Microbiol 6:213–218. doi:10.1016/S1369-5274(03)00060-2
Acknowledgments
This work was funded by the ‘Instituto de Ciencia y Tecnología del Distrito Federal (Mexico)’ project ICyTDF/295/2009. J.M. B.-L., Y.E. N.-N, S. G.-A, and Z. H.-M. received grant-aided support from ‘Consejo Nacional de Ciencia y Tecnología’ (CONACYT, Mexico). Escherichia coli BW20767 carrying plasposon pRL27 and E. coli DH5α/λpir were kindly provided by Dr. William W. Metcalf (University of Illinois at Urbana–Champaign, USA). Escherichia coli S17-1 λpir carrying the plasmid pKNOCK-Cm was provided by Dr. Mikhail F. Alexeyev (University of South Alabama, Mobile, AL, USA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bello-López, J.M., Navarro-Noya, Y.E., Gómez-Acata, S. et al. Identification of α-amylase by random and specific mutagenesis of Texcoconibacillus texcoconensis 13CCT strain isolated from extreme alkaline–saline soil of the former Lake Texcoco (Mexico). Folia Microbiol 59, 235–240 (2014). https://doi.org/10.1007/s12223-013-0289-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-013-0289-8