Abstract
Grifola frondosa (“Maitake”) is an edible fungus with several nutraceutical properties, largely used in traditional medicine. The increased use of Maitake as a food supplements ingredient raised the need of accurate authentication methods since the morphological identification of G. frondosa is not feasible in formulated food supplements. We developed a diagnostic tool based on loop-mediated isothermal AMPlification (LAMP) for the detection of G. frondosa in food supplements. First, a modified CTAB protocol for DNA extraction from food supplements has been set up and it has been shown to be able to isolate amplifiable total genomic material from different types of commercial products. Subsequently, the LAMP assay confirmed high specificity and good analytical sensitivity, allowing to detect up to 0.62 pg of genomic DNA in less than 20 min. Ten related fungal species resulted negative, confirming the specificity of the assay. The presence of Maitake in commercial food supplements was confirmed, except for one, revealing a mislabeling (or a food fraud). This assay proved to be a rapid powerful tool for food authentication purposes and routine inspections at any level of the supply chain of Maitake-based products and it can be used as a model for other quality control assays of fungal food products.
Similar content being viewed by others
Introduction
Mushrooms are higher fungi belonging to Ascomycetes and Basidiomycetes species, able to produce fruiting bodies possessing nutraceutical properties and considered a potential source of novel natural molecules for exploitation in the pharmaceutical industry. Many of these mushrooms are edible and generally show low amount of calories and fats, as well as a high nutritional value (proteins, mineral salts, and vitamins), and for this they are considered ideal for a healthy diet. However, the main characteristic of higher fungi is represented by their richness in bioactive compounds, such as polysaccharides (mainly glucans), terpenoids, phenolic compounds, and lectins, making them nutraceutical valuable (Sharifi-Rad et al. 2020). The world production of mushrooms with nutraceutical value has been estimated to reach $18 billion in the first years of XXI century (Gargano et al. 2017). Currently, the main mushroom producer and consumer is China, covering almost the 75% of the whole-world production and meeting large external and internal market demands (Royse et al. 2017; Ferraro et al. 2020). Due to their putative antioxidant, anticancer, antimicrobial, and immunomodulating properties, mushrooms are included in hundreds of different food supplements called “mushroom pharmaceutical,” successfully commercialized worldwide (Lindequist 2013; Gargano et al. 2017). Recently, the number of health-conscious consumers concerned with health benefits linked to the consumed food products has grown. The COVID 19 pandemic contributed to the rise in awareness about the food supplements with immunomodulatory effects to prevent health issues. The demand for functional foods with nutraceutical effects boosted the mushroom market, including those of fungal-based food supplements. In the next future, the global mushroom market has been predicted to experience a significant expansion because of this rising awareness of a healthy nutritional lifestyle (Veljović and Krstić 2020).
Nowadays, one of the species, which has gained more and more attention and for which market has started to grow exponentially, is the fungus Grifola frondosa (Dicks.) Gray is a Basidiomycetes fungus belonging to the family of Meripilaceae and the order of Polyporales. In nature, as a wood decay agent, this fungus is found to associate to stumps of a wide range of broadleaf trees, and its distribution covers temperate forests in North America, Europe, and Asia (Wu et al. 2021). In several countries, particularly in Asia (e.g., Japan, China), the fungus is cultivated in bottle, bag, and outdoor bed cultures, in order to produce the high value fruiting bodies also know with the japanese name of Maitake and the chinese name “hui-shu-hua” (gray tree flower) (Mayuzumi et al. 1997). In the last years, this mushroom ranked 11th among all cultivated fungal species in the world in terms of annual output (Chang, 2006) and both fresh and dried Maitake basidiomata are commercialized in Asia, EU, USA, and Canada (Ferraro et al. 2020). Good taste and delicate aroma make this fungus as excellent to be consumed fresh; however, due to its peculiar biochemical profile, G. frondosa is not only used as a food ingredient, but also as nutraceutical component in food supplements based on mushrooms, generally in dried powder form (Wu et al. 2021). In fact, G. frondosa fruiting bodies are reported to harbor a wide range of bioactive polysaccharides and fractions, such as the β-glucan D-fraction, a complex with about 30% protein, MD-fraction, X-fraction (1 → 6)-β-glucan with (1 → 4)-α branches, Grifolan, and α-D-glucan (He et al. 2017). The plethora of effects of the different polysaccharide fractions isolated from G. frondosa has been documented, and included immunomodulation, antitumor, antiviral, antidiabetic, and anti-inflammation (He et al. 2017). In recent years, a link between G. frondosa polysaccharides and changes in gut microbiota, which play a role in maintaining immune homeostasis and may be related to antitumoral effects, has been also reported (Liu et al. 2019). Moreover, other compounds extracted from G. frondosa fruiting bodies and mycelia have shown promising medicinal values, such as the protein components, sterols, and phenolic compounds (Wu et al. 2021).
The high economic value, the growing interest, and the increased use of Maitake in food supplements highlighted the need of authentication methods of G. frondosa in mushroom-based products. Food authenticity is currently a subject of great concern to food authorities, since incorrect labeling of foods can represent a commercial fraud and it is subject to sanctions in several countries (Danezis et al. 2016). Moreover, the increased awareness of consumers regarding the composition of foods resulted in a growing interest in the verification of the labeling statements. Hence, it is pivotal to establish that species of high commercial value declared in product labels are not partially or entirely replaced by other species of lower value. This issue has been often reported for cult food based on high value fungal species (e.g., truffles, “porcini”) or yeasts used as starter cultures (Arlorio et al. 1999; Mello et al. 2006; Rizzello et al. 2012). The taxonomic identification of fungal species used in commercial products is therefore a critical need, providing information on samples sold as food supplements, and ensuring quality, correct price value, correct labeling, and food safety (Raja et al. 2017; El Sheikha and Hu, 2018). However, traditional identification methods (e.g., morphology) are not feasible for processed fungi (e.g. dried, milled, lyophilized); also, chemical extraction might disrupt fungal mycelia and associated morphological characteristics (Raja et al. 2017). Molecular methods such as those based on analysis of DNA (e.g., rnd-point/real-time PCR) are now widely and routinely used in food authentication as they are generally efficient, sensitive allowing the fast detection and specific identification of a range of animal, plant, and fungal species in food (Böhme et al. 2019). Recently, DNA-based methods were demonstrated to be also suitable for the authentication of botanical ingredients in herbal food supplements (Grazina et al. 2020). Among them, loop-mediated isothermal amplification (LAMP) is a molecular tool offering fast, accurate, and cost-effective analysis. The LAMP is an isothermal amplification assay which formally works without thermocycler, requiring two sets of primers (external primers F3 and B3, and internal primers, FIP and BIP). The LAMP protocol requires DNA polymerase without exonuclease activity, with strand-displacing activities, in order to generate amplicons containing loop regions to which further primers (namely FL and BL) can bind (Notomi et al. 2000). The LAMP enzyme is rather tolerant to chemical inhibitors often found in environmental samples and complex food matrices compared to standard PCR polymerases, making LAMP assay as feasible for processing DNA extracted from complex raw materials (Tomlinson, 2013) like processed ingredients and foods. The application of LAMP-based assays in screening of food for authentication and safety has started to be popular, and it has been reported for different kinds of food products. Regarding food safety, much attention has been paid to the detection of foodborne pathogens (Zhong and Zhao 2018), genetically modified (GM) food contents (Singh et al. 2019) and allergenic ingredients such as pistachio (Pistacia vera) and peanut (Arachis hypogea) (Sheu et al. 2018; Mao et al. 2020). For the authentication of food products, the LAMP technique has been largely used for the identification of animal species in meat products (chicken, ostrich, buffalo, pork) (Tasrip et al. 2019), but also to discriminate some medicinal plant species (“botanicals”), like Curcuma longa, Panax ginseng, Crocus sativus (saffron), and Hedyotis diffusa, an ingredient of popular herbal teas (Sasaki and Nagumo, 2007; Sasaki et al., 2008; Zhao et al. 2016; Li et al. 2013).
Here, we report on the development of a LAMP diagnostic assay for the detection of G. frondosa in food supplements. The assay has been evaluated for analytical specificity and sensitivity. A DNA extraction method optimized for mushroom-based food supplements has been developed; finally, the assay has been used to assess the presence of G. frondosa in several commercial food supplements with diverse chemical composition and different formulation.
Materials and Methods
Investigated Materials
Fungal Samples
In order to evaluate the sensitivity and the specificity of the LAMP assay, ten different fungal species were used. The target species, Grifola frondosa, was obtained from a fruiting body acquired in a local market, certified by expert mycologists, and by molecular assay (see below). Nine fungal species were selected as non-target controls: one species, Ganoderma carnosum Pat., selected as a closed representative of the Polyporaceae family, was collected in the wild and identified through molecular diagnostic assays. Eight species were selected as they are the most commercialized mushrooms and their carpophores were obtained in a local supermarket: Agaricus bisporus (J.E. Lange) Imbach (champignon), Boletus spp. (porcino), Cyclocybe aegerita (V. Brig.), Vizzini (poplar mushroom), Lentinula edodes (Berk.), Pegler (Shitake), Pleurotus ostreatus (Jacq.), P. Kumm. (oyster mushroom), Saccharomyces cerevisiae (Desm.) Meyen (yeast). Finally, two species of the most common fungi widely distributed in soil and indoor environments were selected, Fusarium sp. and Cladosporium sp. The two isolates were obtained from agar plates filled with MEA left open for 24 h, in an indoor environment. Isolates were identified as Fusarium equiseti and Cladosporium ramotenellum by molecular approach outlined below, according to the closest sequences present in NCBI database.
Food Supplements Containing Mushroom
Different commercial food supplements containing fungal ingredients were selected to be tested with the developed assay. Supplements were both commercial complex formulation and powdered mushroom blends. These products were acquired at local or online pharmacies and stores. Names, declared quality/quantity composition and formulation (powder, capsules or compression tablets), concerning each sample, are listed in Table 1.
Genomic DNA extraction
Total genomic DNA extraction from all fungal samples as well as from commercial products was performed following the CTAB method described by Doyle and Doyle (1990) with some modifications. Briefly, powdered materials obtained from fungal carpophores and mycelia (approximately 20 mg), fungal supplements (60–80 mg), or dried-powdered fungal carpophores (20 mg) were mixed with 1 mL of CTAB extraction buffer (2% of CTAB (w/v), 0.1 mol L−1 of TrisHCl, 1.4 mol L−1 of NaCl, 0.02 mol L−1 of EDTA, pH 8.0), previously added with 2% PVP (w/v) and 1% PVPP (w/v). After the addition of 2% of β-mercaptoethanol, the mixture was incubated at 65 °C for 1 h in a thermal block under continuous stirring (400 rpm).
All samples were firstly purified by liquid–liquid extraction with phenol/chloroform/isoamyl alcohol (25:24:1 v/v) and later with chloroform/isoamyl alcohol (24:1 v/v) two times. DNA precipitation was performed with 0.7 volume parts of isopropanol and the solution was incubated for 1 h at 4 °C. DNA was dissolved in 50 µL of distilled water and all extracts were immediately kept at − 20 °C until further analysis. DNA concentration and purity were assessed using a NanoPhotometer® P-Class (Implen).
LAMP Primers Design
The primers set for the LAMP assay was developed using the PrimerExplorer V5 software (http://primerexplorer.jp/lampv5e/index.html), and the internal transcribed spacers of nuclear ribosomal DNA (ITS1-5.8S-ITS2) sequence was selected as a target region (hereafter referred as “ITS”). To obtain a specific region for Grifola frondosa, ITS sequences of this fungus (see below) were compared to 66 ITS sequences from closely related non-target species obtained from the National Center for Biotechnology Information (NCBI) GenBank database (Table S1). Fungal species belonging to the Polyporaceae family and closely related to G. frondosa have been identified thanks to the phylogenetic study of Justo et al. (2017). Six available ITS sequences of different isolates of G. frondosa (GBPOL1291-13, GBPOL2259-14, GBPOL2668-15, GBPOL320-13, NOBAS1417-15, NOBAS1859-16) were also selected from the Barcode of Life (BOLD) database (https://www.boldsystems.org/index.php/databases) to avoid intraspecific nucleotide diversity that could prevent an accurate detection of the target species. All selected sequences were aligned using the software MEGA X (Kumar et al. 2018) using ClustalW algorithm and a small region (222 bp) showing high nucleotide diversity between species and no intraspecific diversity was selected for the primers design. A set of five oligonucleotide primers, including two outer primers (forward primer F3 and backward primer B3), two inner primers (forward inner primer FIP and backward inner primer BIP), and only one loop primer (forward loop primer LoopF) were developed following the standard parameter sets of the software. The key factors in the LAMP primer design are the Tm (about 65 °C for each region), the stability at the end of each primer, the GC content (between about 40 and 65%), and secondary structure. The distance between the different primers is also important: the region amplified by the LAMP method is considered to be between 120 and 180 bases, the distance for loop forming regions is 40–60 bp and the distance between F2 and F3 (and B2 and B3) is between 0 to 60 bases. Primers were synthesized by Metabion international AG and the sequences are listed in Table 2.
PCR Assays
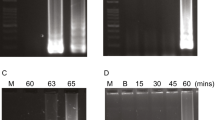
Qualitative PCR amplification was performed to each DNA (i) to evaluate the suitability for amplification, particularly for DNA from food supplements, and (ii) for confirming the identity of the fungal species selected for the specificity test. The reaction was carried out in 25 µL of total volume containing 1.5 µL of DNA isolated from fungal carpophores or food supplements, 1 × PCR Mix Plus (A&A Biotechnology) and 0.5 µM of each primer ITS1F–ITS2 (White et al. 1990; Gardes and Bruns 1993). PCR reactions were carried out in a T100 Thermal cycler (Bio-Rad, Milan, Italy) using the following program: denaturation step of 95 °C for 3 min, 40 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C of 45 s, and a final extension at 72 °C for 5 min. The amplified products were then analyzed by electrophoresis on 1.5% TBE agarose gel (Bio-Rad, Milan, Italy), stained by Atlas ClearSight DNA Stain (Bioatlas, Tartu, Estonia). The agarose gel was visualized under UV light using Multimage II-AlphaImager® HP (Alpha Innotech, CA, USA) and digital image was obtained with Alpha View—AlphaImager® HP software.
Real-Time LAMP Assay
LAMP reactions were performed in a total volume of 25 µL containing 0.25 µL of each external primers F3 and B3 (200 nM), 0.25 µL of each internal primers FIP and BIP (500 nM), 0.125 µL of the loop primer FL (500 nM), 15 μL of Isothermal Mastermix ISO-001 (OptiGene, UK), and 1 μL of fungal DNA template or 1.5 μL of commercial products DNA. LAMP assays were carried out using an ABI PRISM 7900 HT Sequence Detection System (Life Technologies, Monza, Italy) equipped with fluorescein (FAM) reading channel. All reactions were carried out at 65 °C for 40 min (with a read plate every minute) with a melting curve analysis step (ramp from 80 to 95 °C with a temperature increment of 0.1 °C and a read plate every 10 s).
The LAMP products were visualized as amplification curves in a plot where the Ct value of 1 corresponds to one min.
G. frondosa LAMP Primers Specificity Assay
The genomic DNA extracted from nine different fungal species, including the target one (G. frondosa), and non-target species, was used in the LAMP assay for specificity determination. DNA amplifiability of non-target samples was first assessed in PCR using ITS1F-ITS2 primers, following the procedure described above (section “PCR Assays”). For LAMP reactions, about 10 ng of DNA for each target and non-target species was used as template. Two technical replicates were performed for each biological sample.
Sensitivity Assessment of the LAMP Assay
The sensitivity of the LAMP assay was determined using a tenfold serial dilution of the genomic DNA isolated from a G. frondosa carpophore, ranging from 25 ng/μL to 25 × 10−5 ng/μL. Five technical replicates were tested for each dilution. To simulate the influence of the matrix effect in food supplements, the sensitivity of the assay was also assessed by diluting the genomic DNA of G. frondosa in an extract of mushrooms commercial product without Maitake between its ingredients instead of water, using a tenfold serial dilution as above. The LOD, calculated according to Shrivastava and Gupta (2011), was defined as follows: LOD = 3.3 × (SD/s), where SD is the standard deviation of the y intercept and s is the slope of the curve. The coefficient 3.3 was used assuming a 95% confidence level (Shrivastava and Gupta 2011).
Purification, Sequencing and BLAST Analysis of PCR Products
In order to confirm the identity of fungal samples used in this study and to verify that the PCR products of the LAMP assay corresponded to the target species G. frondosa, Sanger sequencing of amplicons was performed. For this purpose, all the DNA samples were initially amplified using both the ITS1F/ITS2 and ITS1F/ITS4 primers. However, the primers pair with ITS4 (those usually used for the DNA barcoding of the ITS region in fungi) was unable to amplify the DNA isolated from the food supplements, probably depending on the partial degradation of the fungal DNA correlated to processing and on the “matrix effect” (excipients). So, only the PCR products obtained using ITS1F-ITS2 primers and with the external LAMP primers F3-B3 were analyzed, after being purified using the Wizard® SV Gel (Promega Corp., Madison, WI, USA) and PCR Clean-Up System (Promega) following the manufacturer’s instructions. The purified DNA was eluted in 30 μL of nuclease free water and Sanger sequencing of the purified PCR products was performed at Sequencing Service of the Genomics Service Unit (Munich) using BigDye Terminator v3.1 cycle sequencing. The sequences were obtained bidirectionally using both strands with a combination of the following primers: ITS1F (Gardes and Bruns 1993)/ITS1 and ITS2. For both sequencing reactions, approximately 5 μL of PCR template was used along with 3-μM sequencing primers. Sequences were assembled with Sequencher 5.2.3 (Gene Codes, Ann Arbor, MI, USA), optimized, and then corrected manually when necessary; the latter step was performed to ensure that proper base calls were assigned by the algorithm. Each sequence was subjected to an individual Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990) search to verify nucleotide identity against a sequence database. BLASTn search was employed using nucleotide collection (nr/nt), excluding uncultured/environmental sample sequences.
Results
Development of Specific LAMP Primers for the G. frondosa Species
The multiple alignment of G. frondosa ITS1-5.8S-ITS2 nuclear ribosomal DNA sequences with the same genomic regions of other related fungal species allowed us to identify a region of 222 nucleotides with high interspecific variation and no intraspecific variation within the isolates of the target species. On this specific region of G. frondosa sequence, a set of LAMP primers were developed as shown in Fig. 1.
Multiple alignment between the internal transcribed spacer (ITS) of G. frondosa and related species for the design of the specific LAMP primer sets. In the boxes are represented the six loci (F3, F2, F1, B3, B2, B1) of G. frondosa ITS where the primers are located. The identity between the ITS sequences of the selected species is indicated with a green bar and polymorphic sites are highlighted in colors. The arrows indicate the direction of the LAMP amplification
In order to evaluate the specificity of the new set of primers, DNA samples isolated from ten different fungal species, including G. frondosa, were used for the LAMP assay (Fig. 2a). Only the DNA of the target species G. frondosa was successfully amplified with the newly developed LAMP primers, confirming the specificity of the assay. No amplification curves were detected for all the nine non-target species (Fig. 2b) and melting curve analysis confirmed the absence of non-specific amplicons, both for non-target species and the negative control (Fig. 3).
Specificity of the LAMP primers for the identification of G. frondosa. a Amplicons obtained from genomic DNA extracted from carpophores of ten fungal species analyzed with ITS1-2 primers. b Plot showing the real-time amplification curve of the positive control (G. frondosa carpophore DNA), while no amplification is visible for non-target species (A. bisporus, Boletus spp., Cladosporium sp., C. cylindracea, Fusarium sp., G. carnosum, L. edodes, P. ostreatus, S. cerevisiae)
Analytical Sensitivity and Limit of Detection (LOD) of the LAMP Assay
In order to calculate the analytical sensitivity of the LAMP assay, ten-fold serial dilutions were prepared starting from genomic DNA extracted from a G. frondosa carpophore (positive control) and were amplified with the LAMP primers. Results showed that G. frondosa detection time ranged from 7 to 13 min (Fig. 4a). The limit of detection (LOD) of the assay resulted 0.62 pg/μL.
Sensitivity of the LAMP assay for G. frondosa DNA. Amplification curves of a serial tenfold dilution of the genomic DNA extracted from a carpophore of G. frondosa (from 25 ng/µL to 0.2 pg/µL). The target DNA was analyzed diluted in water (plot a) and in a supplement extract (plot b) to simulate the matrix effect of a food supplement
To estimate the analytical sensitivity of the G. frondosa LAMP assay in commercial food supplements, the LOD was also calculated using target DNA diluted in a supplement extract. In this case, our method allowed the detection of about 2.5 pg of target DNA with a time of detection of 16 min (Fig. 4b). The LOD of the assay on DNA diluted in a commercial food supplement extract was slightly higher (0.70 pg/μL) compared to LOD resulted from sensitivity assay on water. Standard curves based on time of detection (Ct) and DNA concentration (pg) of analytical sensitivity assay on water and commercial food supplements showed R2 of 0.99 and 0.95, respectively (Fig. 5).
Application of the LAMP Assay for the Detection of G. frondosa in Mushrooms Food Supplements
Ten different commercial products containing G. frondosa were selected and purchased to assess if the LAMP assay can be suitable for the detection and the authentication of food supplements containing Maitake mushrooms. Various types of commercial products were selected to evaluate the impact of different processing steps and formulations (especially pre-processing like drying or lyophilization, the composition of excipients as well as the presence of DNA of other fungal species) on the isolation of a DNA with sufficient quality to allow downstream applications. The CTAB modified method used in this study for the DNA extraction resulted to be a suitable protocol for the isolation of nucleic acids from mushroom-based food supplements. Prior to the LAMP amplification, the total genomic DNA extracted from all types of commercial products was tested in traditional PCR with the ITS1F-ITS2 primer set. All the DNA samples were successfully amplified with this primer set (Figure S1). Once the amplifiability of the DNA has been ascertained, the LAMP assay was performed on all samples. Using the ITS-based LAMP primers developed herein, G. frondosa was detected in nine out of ten commercial products that were labeled to contain Maitake, confirming also the effectiveness of the DNA extraction method. An amplification plot for each type of mushroom product (capsule, powder, tablet, and chewable tablet) was created, including a positive control (G. frondosa DNA) (Fig. 6). Regarding the first type of supplements, the capsules, all analyzed products showed a positive signal in the LAMP assay within a time of detection of less than 20 min, except for the sample I2 that resulted as negative (Fig. 6a). All the samples in powder showed an amplification signal ranging from 8.8 to 35.3 min, with the sample I9 showing a time of detection of 35.3 min ± 0.85 (Fig. 6b). Finally, all tablet samples showed positive signals: the tablet (I6) showed a very low time of detection, around 10 min (Fig. 6c), while the chewable tablet (I8) showed a time of detection around 30 min (Fig. 6d). Sequencing of some representative amplicons (sample I6 and I7) confirmed that the positive signal obtained in food supplement samples belong to G. frondosa, after blastn analysis. The sequence obtained from the sequencing of ITS region from I2 sample, resulting negative to LAMP assay, showed the best match (100% nucleotide identity, e-value 4e−91, query coverage 95%) with a sequence of a Boletus sp. isolate (accession number AB821462.1).
Application of LAMP assay for the detection of G. frondosa in mushroom food supplements. An amplification plot was generated for each type of supplement: a capsule—I1, I2, I3, I7, I10; b powder—I4, I5, I9; c tablet—I6; d chewable tablet—I8. G. frondosa DNA isolated from a carpophore was used as positive control in each plot. NC, negative control
Discussion
Thanks to their richness in bioactive compounds and their traditional use in Asian medicine, fungi are increasingly used for health purposes, namely in food supplements area, and they have become an important source of bioactive compounds and nutraceuticals (Cateni et al., 2021). As the market for mushroom-based products grows, so does the need for more rapid and accurate controls to prevent fraud. Little attention has been put on this problem and very few works take advantage of molecular, DNA-based techniques to authenticate nutraceuticals containing mushrooms (Loyd et al., 2018; Raja et al., 2017; Zhang et al., 2020).
The goal of this study was the set-up and validation of a rapid and specific LAMP assay, coupled with an optimized protocol for DNA extraction from food supplements, aimed to quickly detect and authenticate Maitake mushroom in fungal-based food supplements.
Extraction of DNA from these complex products, in which either mycelium and fruiting bodies of mushrooms are present in the form of extracts or dried biomass, was successfully achieved by the optimization of a CTAB-based method. It should be highlighted that the treatments to which the mushrooms are subjected to prepare the final product (food supplement), as well as the presence of excipients, might significantly affect both quantity and integrity of fungal DNA, and its subsequent amplification. The CTAB modified method used in this work was demonstrated to be effective in the extraction of DNA from all the different type of mushroom-based products. Compared to the commercially available DNA purification kits, the protocol allowed the processing of a larger quantity of starting sample (up to 80 mg), resulting in a satisfying extraction yield. As demonstrated by Lu et al. (2018), the DNA extracted from botanical extracts is often fragmented (fragments of less than 300 bp), implying that DNA amplifications were possible only with the use of primer pairs producing small amplicons (such as the ITS1F-ITS2 primer couple, designed on the repeated ribosomal genomic DNA). This important aspect was taken into account for the selection of the G. frondosa species-specific locus prior to LAMP primers design, and a short region (222 bp) of the ITS sequence was used for the purpose. Internal Transcribed Spacer (ITS) region of nuclear DNA was selected as candidate locus because it is one of the recognized universal barcode markers for fungal kingdom and has become the most sequenced region to identify fungal taxonomy at species level (Schoch and Seifert 2012). Our results showed that the newly designed set of ITS-based primers allows a specific identification of Maitake DNA in commercial products based on fungal ingredients. Both the forward primers (F3 and FIP) and the backward ones (BIP and B3) were designed to cover a region with high number of polymorphic sites between species, allowing specific detection of the target species (G. frondosa) and avoiding any potential cross reaction with nine selected fungal species, as demonstrated by the specificity assay. The LAMP assay coupled with real-time PCR detection system allowed to obtain clear positive amplification results for the target control G. frondosa in less than 20 min. Our data showed that the minimum concentration of target DNA detected by the LAMP assay was 0.62 pg/μL and that a putative matrix effect due to the composition of a supplement (excipients and presence of other species) could affect negatively the sensitivity of the assay only to a small extent (0.70 pg/μL). As already demonstrated in several studies, the LAMP technique, compared with the conventional PCR assay, shows a higher sensitivity and is more tolerant to well-known PCR inhibitors, demonstrating to be an ideal molecular technique for identifying botanical ingredients (Zhao et al., 2016; Mao et al., 2020). In Table 3, we summarize the pros and cons of both LAMP and traditional DNA barcoding of ITS region.
In this study, we have successfully developed and applied LAMP technique for the identification of Maitake ingredient in different types of commercial food supplements. In nine out of ten products, it was possible to detect the DNA of G. frondosa, confirming what was declared on the label. The time of the detection varied according to the type of treatment that the fungus has undergone: products containing Maitake-dried carpophore were detected earlier than those with fungal extract, and lyophilized mushrooms showed a time of detection lower than mushrooms subjected to other types of drying. The most difficult products to analyze were those containing excipients and composed from different fungal species (I3, I8, I9). In these, the DNA of G. frondosa was successfully detected beyond 30 min. Only one sample (I2), a food supplement labeled to contain Maitake as the only fungal ingredient, resulted as negative after the LAMP assay. Sequencing of ITS sequence amplified from DNA of sample I2 revealed that the amplified fungal DNA belong to Boletus spp., an edible cultivated fungus, and more common (at least in Europe, where the food supplements I2 was produced) than G. frondosa. According to our LAMP assay and the sequencing of fungal DNA of the sample, G. frondosa DNA resulted absent in the I2 sample; however, we cannot exclude a presence of G. frondosa DNA below the LOD of the assay (0.62 pg). On the other hand, the presence of Boletus spp. suggests that this fungus replaced G. frondosa as main ingredient of I2 sample, or that the fungus contaminated the sample during transformation processes (e.g., during milling). Others authors have shown that DNA-based approaches can be useful to detect species substitutions in consumer products containing fungi: for example, Raja et al. (2017) found that many of the products labeled with Cordyceps sinensis and Ganoderma lucidum (Reishi) contained other fungal species. These studies confirm the usefulness of the molecular analysis in order to detect and fight food frauds in food supplements area, or simply to detect involuntary mislabeling and cross-contamination of fungal food ingredients. In fact, our results demonstrated that LAMP is an efficient tool to detect mushroom ingredients in food supplements and a novel molecular technique is provided for the rapid identification of G. frondosa, the famous Maitake mushroom. Given the importance of the correct use of ingredients for the quality assurance of food and food supplements, this strategy—which combines the speed and efficiency of LAMP and the accuracy of ITS barcode—may be extended to other nutraceutical products that need to be certified at the species level, and it can be used for routine inspections at any level of the production chain. In addition, as already demonstrated in plant pathogen studies (Sillo et al. 2018), LAMP assay can be also performed using in-field portable equipment, such as the Genie® II from OptiGene, becoming a possible practical solution for surveillance and monitoring throughout the food production chain also in laboratories not equipped with qPCR instruments.
Conclusions
In the present study, a LAMP assay based on the ITS gene was developed for the specific, sensitive, and rapid identification of Grifola frondosa, also known as Maitake mushroom. With the developed LAMP primer set, less than 1 pg of G. frondosa DNA could be detected without cross-amplification. This novel molecular approach can be applied for heat-processed Maitake products and for commercial food supplements to perform their authentication and prevent food fraud and adulteration.
Data Availability
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Arlorio M, Coisson JD, Martelli A (1999) Identification of Saccharomyces cerevisiae in bakery products by PCR amplification of the ITS region of ribosomal DNA. Eur Food Res Technol 209(3):185–191. https://doi.org/10.1007/s002170050477
Böhme K, Calo-Mata P, Barros-Velázquez J, Ortea I (2019) Review of recent DNA-based methods for main food-authentication topics. J Agric Food Chem 67(14):3854–3864. https://doi.org/10.1021/acs.jafc.8b07016
Cateni F, Gargano ML, Procida G, Venturella G, Cirlincione F, Ferraro V (2021). Mycochemicals in wild and cultivated mushrooms: nutrition and health. Phytochem Rev, 1-45. https://doi.org/10.1007/s11101-021-09748-2
Chang ST (2006) The world mushroom industry: trends and technological development. Int J Med Mushrooms 8(4):297–314. https://doi.org/10.1615/IntJMedMushr.v8.i4.10
Danezis GP, Tsagkaris AS, Brusic V, Georgiou CA (2016) Food authentication: state of the art and prospects. Curr Opin Food Sci 10:22–31. https://doi.org/10.1016/j.cofs.2016.07.003
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12(13):39–40. https://doi.org/10.1007/978-3-642-83962-7_18
El Sheikha AF, Hu DM (2018) How to trace the geographic origin of mushrooms? Trends Food Sci Technol 78:292–303. https://doi.org/10.1016/j.tifs.2018.06.008
Ferraro V, Venturella G, Pecoraro L, Gao W, Gargano ML (2020). Cultivated mushrooms: importance of a multipurpose crop, with special focus on Italian fungiculture. Plant Biosyst, 1-11. https://doi.org/10.1080/11263504.2020.1837283
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
Gargano ML, van Griensven LJ, Isikhuemhen OS, Lindequist U, Venturella G, Wasser SP, Zervakis GI (2017) Medicinal mushrooms: valuable biological resources of high exploitation potential. Plant Biosyst 151(3):548–565. https://doi.org/10.1080/11263504.2017.1301590
Grazina L, Amaral JS, Mafra I (2020) Botanical origin authentication of dietary supplements by DNA-based approaches. Compr Rev Food Sci Food Saf 19(3):1080–1109. https://doi.org/10.1111/1541-4337.12551
He X, Wang X, Fang J, Chang Y, Ning N, Guo H, Huang L, Huang X, Zhao Z (2017) Polysaccharides in Grifola frondosa mushroom and their health promoting properties: a review. Int J Biol Macromol 101:910–921. https://doi.org/10.1016/j.ijbiomac.2017.03.177
Justo A, Miettinen O, Floudas D, Ortiz-Santana B, Sjökvist E, Lindner D, Nakasone K, Niemelä T, Larsson KH, Ryvarden L, Hibbett DS (2017) A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol 121(9):798–824. https://doi.org/10.1016/j.funbio.2017.05.010
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547. https://doi.org/10.1093/molbev/msy096
Li M, Wong YL, Jiang LL, Wong KL, Wong YT, Bik-San Lau C, Shaw PC (2013) Application of novel loop-mediated isothermal amplification (LAMP) for rapid authentication of the herbal tea ingredient Hedyotis diffusa Willd. Food Chem 141(3):2522–2525. https://doi.org/10.1016/j.foodchem.2013.05.085
Lindequist U (2013) The merit of medicinal mushrooms from a pharmaceutical point of view. Int J Med Mushrooms 15(6):517–523. https://doi.org/10.1615/IntJMedMushr.v15.i6.10
Liu L, Li M, Yu M, Shen M, Wang Q, Yu Y, Xie J (2019) Natural polysaccharides exhibit anti-tumor activity by targeting gut microbiota. Int J Biol Macromol 121:743–751. https://doi.org/10.1016/j.ijbiomac.2018.10.083
Loyd AL, Richter BS, Jusino MA, Truong C, Smith ME, Blanchette RA, Smith JA (2018) Identifying the “Mushroom of Immortality”: assessing the Ganoderma species composition in commercial Reishi products. Front Microbiol 9:1557. https://doi.org/10.3389/fmicb.2018.0155
Lu Z, Rubinsky M, Babajanian S, Zhang Y, Chang P, Swanson G (2018) Visualization of DNA in highly processed botanical materials. Food Chem 245:1042–1051. https://doi.org/10.1016/j.foodchem.2017.11.067
Mao R, Xie K, Zhao M, Li M, Lu L, Liu Y, Wu Q, Chen Y, Zhang T, Diao E (2020) Development and evaluation of a loop-mediated isothermal amplification (LAMP) assay for rapid detection of pistachio (Pistacia vera) in food samples. Food Anal Methods 13(3):658–666. https://doi.org/10.1007/s12161-019-01684-4
Mayuzumi Y, Mizuno T (1997) Cultivation methods of maitake (Grifola frondosa). Food Rev 13:357–364. https://doi.org/10.1080/87559129709541117
Mello A, Ghignone S, Vizzini A, Sechi C, Ruiu P, Bonfante P (2006) ITS primers for the identification of marketable boletes. J Biotechnol 121(3):318–329. https://doi.org/10.1016/j.jbiotec.2005.08.022
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12):e63–e63. https://doi.org/10.1093/nar/28.12.e63
Raja HA, Baker TR, Little JG, Oberlies NH (2017) DNA barcoding for identification of consumer-relevant mushrooms: a partial solution for product certification? Food Chem 214:383–392. https://doi.org/10.1016/j.foodchem.2016.07.052
Rizzello R, Zampieri E, Vizzini A, Autino A, Cresti M, Bonfante P, Mello A (2012) Authentication of prized white and black truffles in processed products using quantitative real-time PCR. Food Res Int 48(2):792–797. https://doi.org/10.1016/j.foodres.2012.06.019
Royse DJ, Baars J, Tan Q (2017) Current overview of mushroom production in the world. In: Zied DC, Pardo-Giminez A (eds) Edible and medicinal mushrooms: technology and applications. New York, John Wiley & Sons LtD, Hoboken, pp 5–13. https://doi.org/10.1002/9781119149446.ch2
Sasaki Y, Nagumo S (2007) Rapid identification of Curcuma longa and C aromatica by LAMP. Biol Pharm Bull 30(11):2229–2230. https://doi.org/10.1248/bpb.30.2229
Sasaki Y, Komatsu K, Nagumo S (2008) Rapid detection of Panax ginseng by loop-mediated isothermal amplification and its application to authentication of ginseng. Biol Pharm Bull 31(9):1806–1808. https://doi.org/10.1248/bpb.31.1806
Schoch C, Seifert KA (2012) Reply to Kiss: Internal transcribed spacer (ITS) remains the best candidate as a universal DNA barcode marker for fungi despite imperfections. PNAS 109(27):E1812–E1812. https://doi.org/10.1073/pnas.1207508109
Sharifi-Rad J, Butnariu M, Ezzat SM, Adetunji CO, Imran M, Sobhani SR, Tufail T, Hosseinabadi T, Ramírez-Alarcón K, Martorell M, Maroyi A, Martins N (2020) Mushrooms-rich preparations on wound healing: from nutritional to medicinal attributes. Front Pharmacol 11:1452. https://doi.org/10.3389/fphar.2020.567518
Sheu SC, Tsou PC, Lien YY, Lee MS (2018) Development of loop-mediated isothermal amplification (LAMP) assays for the rapid detection of allergic peanut in processed food. Food Chem 257:67–74. https://doi.org/10.1016/j.foodchem.2018.02.124
Shrivastava A, Gupta V (2011) Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron Young Sci 2(1):21–21
Sillo F, Giordano L, Gonthier P (2018) Fast and specific detection of the invasive forest pathogen Heterobasidion irregulare through a loop-mediated isothermal AMP lification (LAMP) assay. For Pathol 48(2):e12396. https://doi.org/10.1111/efp.12396
Singh M, Pal D, Sood P, Randhawa G (2019) Loop-mediated isothermal amplification assays: rapid and efficient diagnostics for genetically modified crops. Food Control 106:106759. https://doi.org/10.1016/j.foodcont.2019.106759
Tasrip NA, Khairil Mokhtar NF, Hanapi UK, Abdul Manaf YN, Ali ME, Cheah YK, Mustafa S, Mohd Desa MN (2019) Loop mediated isothermal amplification; a review on its application and strategy in animal species authentication of meat-based food products. Int Food Res J 26(1):1–10
Tomlinson J (2013). In-field diagnostics using loop-mediated isothermal amplification. In M. Dickinson, & J. Hodgetts (Eds.), Phytoplasma: methods and protocols, Vol. 938 (pp. 291–300). New York, NY: Humana Press. https://doi.org/10.1007/978-1-62703-089-2_25
Veljović S, Krstić J (2020) Elaborating on the potential for mushroom-based product market expansion: consumers’ attitudes and purchasing intentions. In: Singh J, Meshram V, Gupta M (eds) Bioactive natural products in drug discovery. Springer, Singapore, pp. 643–663. https://doi.org/10.1007/978-981-15-1394-7_23
White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, United States, pp 315–322
Wu JY, Siu KC, Geng P (2021) Bioactive ingredients and medicinal values of Grifola frondosa (Maitake). Foods 10(1):95. https://doi.org/10.3390/foods10010095
Zhang FL, Yang XF, Wang D, Lei SR, Guo LA, Liu WJ, Song J (2020) A simple and effective method to discern the true commercial Chinese cordyceps from counterfeits. Sci Rep 10(1):1–8. https://doi.org/10.1038/s41598-020-59900-9
Zhao M, Yuhua S, Lan W, Licheng G, Wei L, Chao X, Song Y, Wei S, Shilin C (2016) Rapid authentication of the precious herb saffron by loop-mediated isothermal amplification (LAMP) based on internal transcribed spacer 2 (ITS2) sequence. Sci Rep 6:25370. https://doi.org/10.1038/srep25370
Zhong J, Zhao X (2018) Isothermal amplification technologies for the detection of foodborne pathogens. Food Anal Methods 11(6):1543–1560. https://doi.org/10.1007/s12161-018-1177-2
Acknowledgements
The authors thank Francesco Venice (IPSP-CNR Torino, Italy) for kindly providing fungal fruiting bodies used in this study and for technical help in fungal isolation and Maria Teresa Gatto for the help in laboratory given during her thesis period.
Funding
Research funded by Regione Piemonte, P.O.R. FESR 2014/2020 (Asse I—Azione I-1b.2.2). “NUTRAcore: Integrated platform for the development of innovative processes in the context of the bio-economy aimed at the sustainable production of functional and safe ingredients for food and nutraceuticals”—Bioeconomy Platform.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Valeria Fochi: data curation, formal analysis, investigation, methodology, writing—original draft. Fabiano Sillo: methodology, resources, formal analysis, writing—original draft. Fabiano Travaglia: resources; Jean Daniel Coïsson: supervision, funding acquisition; Raffaella Balestrini: writing—review and editing; Marco Arlorio: supervision, project administration, funding acquisition, writing—review and editing.
Corresponding author
Ethics declarations
Conflict of Interest
Valeria Fochi declares that she has no conflict of interest. Fabiano Travaglia declares that he has no conflict of interest. Jean Daniel Coïsson declares that he has no conflict of interest. Raffaella Balestrini declares that she has no conflict of interest. Marco Arlorio declares that he has no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fochi, V., Sillo, F., Travaglia, F. et al. A Rapid and Efficient Loop-mediated Isothermal Amplification (LAMP) Assay for the Authentication of Food Supplements Based on Maitake (Grifola Frondosa). Food Anal. Methods 15, 1803–1815 (2022). https://doi.org/10.1007/s12161-022-02235-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-022-02235-0