Abstract
The SARS-CoV-2 (SCoV-2) virus is the causative agent of the ongoing COVID-19 pandemic. It contains a positive sense single-stranded RNA genome and belongs to the genus of Betacoronaviruses. The 5′- and 3′-genomic ends of the 30 kb SCoV-2 genome are potential antiviral drug targets. Major parts of these sequences are highly conserved among Betacoronaviruses and contain cis-acting RNA elements that affect RNA translation and replication. The 31 nucleotide (nt) long highly conserved stem-loop 5a (SL5a) is located within the 5′-untranslated region (5′-UTR) important for viral replication. SL5a features a U-rich asymmetric bulge and is capped with a 5′-UUUCGU-3′ hexaloop, which is also found in stem-loop 5b (SL5b). We herein report the extensive 1H, 13C and 15N resonance assignment of SL5a as basis for in-depth structural studies by solution NMR spectroscopy.
Similar content being viewed by others
Biological context
SCoV-2 is a member of the Betacoronavirus family and contains a large single-stranded (+) RNA genome with a length of approx. 30,000 nucleotides (nts) (Hu et al. 2020; V’kovski et al. 2020). The RNA genome of the virus not only contains the coding regions for the viral proteins, but also extended and highly structured 5′- and 3′-UTRs, as well as internal structured RNA elements with important functional roles in genome replication, transcription of subgenomic (sg) mRNAs and the balanced translation of viral proteins (Madhugiri et al. 2016; Kelly et al. 2020; Tidu et al. 2020). While the development of antiviral therapeutics against COVID-19 is primarily focused on the viral proteins, the highly structured RNA elements provide an extensive reservoir of additional drug targets to be exploited. The architecture of the RNA genome of SCoV2 and related viruses has so far been investigated mainly by sequence-based computational predictions and by chemical probing approaches in vitro and in vivo (e.g. Manfredonia et al. 2020; Rangan et al. 2020). Although structural probing methods have been established to map RNA-small molecule interactions even in cells (Martin et al. 2019), these tools are unable to define the tertiary structure and dynamics of the RNA-elements in the SCoV-2 genome with sufficiently high resolution to enable structure-based drug design by virtual screening.
While the sequences of the individual structural elements vary between different Coronaviruses, their ubiquitous presence and highly conserved secondary structures suggest that these elements are critically important for viral viability and pathogenesis (reviewed in Madhugiri et al. 2016). One example of such an important structure is stem-loop 5 (SL5). SL5 is structurally conserved in the genomes of Alpha- and Betacoronaviruses and has been shown to be crucial for efficient viral replication (Chen and Olsthoorn 2010; Guan et al. 2011).
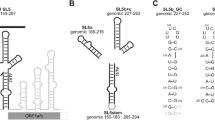
In SCoV-2, SL5 consists of four helices including nts 149–297 of the 5′-UTR and the first 29 nts of the Nsp1 coding region (Suppl. Figure 1A). Sub-elements are joined to the SL5 basal stem by a four-helix junction. These sub-elements are termed SLs 5a, 5b and 5c. SL5a consists of 31 nucleotides and represents the largest of the three stem-loops. Intriguingly, the apical loop sequences of SL5a and SL5b are identical (5′-UUUCGU-3′) and belong to the 5′-UUYCGU-3′ motif, which is also found in Alphacoronaviruses. This high level of sequence conservation suggests functional importance, e.g. in viral packaging (Masters 2019). Thus, we have recently obtained secondary structure models of SL5a-c and the basal stem segment of SL5 based on initial 1H and 15N assignments (Wacker et al. 2020). In order to characterize SL5a further, we provide here a near complete 1H, 13C and 15N chemical shift assignment.
Methods and experiments
Sample preparation
RNA synthesis for NMR experiments: For DNA template production, the sequence of SL5a together with the T7 promoter was generated by hybridization of complementary oligonucleotides and introduced into the EcoRI and NcoI sites of an HDV ribozyme encoding plasmid (Schürer et al. 2002), based on the pSP64 vector (Promega). RNAs were transcribed as HDV ribozyme fusions to obtain a homogeneous 3′-end. The recombinant vector pHDV-5_SL5a was transformed and amplified in the Escherichia coli strain DH5α. Plasmid-DNA was purified using a large scale DNA isolation kit (Gigaprep; Qiagen) according to the manufacturer’s instructions and linearized with HindIII prior to in-vitro transcription using the T7 RNA polymerase P266L mutant, which was prepared as described in (Guillerez et al. 2005). 15 ml transcription reactions [20 mM DTT, 2 mM spermidine, 200 ng/µl template, 200 mM Tris/glutamate (pH 8.1), 40 mM Mg(OAc)2, 12 mM NTPs, 32 µg/ml T7 RNA Polymerase, 20% DMSO] were performed to obtain sufficient amounts of SL5a RNA (5′-pppGGGCUGCUUACGGUUUCGUCCGUGUUGCAGCCC-3′). Preparative transcription reactions (6 h at 37 °C and 70 rpm) were terminated by addition of 150 mM EDTA. SL5a RNA was purified as follows: RNAs were precipitated with one sample volume of ice-cold 2-propanol. RNA fragments were separated on 15% denaturing polyacrylamide (PAA) gels and visualized by UV shadowing at 254 nm. SL5a RNA was excised from the gel and eluted using the following protocol: The gel fragments were granulated in two gel volumes 0.3 M NaOAc solution, incubated for 30 min at − 80 °C, followed by 15 min at 65 °C. The RNA was further eluted from gel fragments overnight by passive diffusion into 0.3 M NaOAc, precipitated with EtOH and desalted via PD10 columns (GE Healthcare). Residual PAA was removed by reversed-phase HPLC using a Kromasil RP 18 column and a gradient of 0–40% 0.1 M acetonitrile/triethylammonium acetate. After freeze-drying of RNA-containing fractions and cation exchange by LiClO4 precipitation (2% in acetone), the RNA was folded in water by heating to 80 °C followed by rapid cooling on ice. Buffer exchange to NMR buffer (25 mM potassium phosphate buffer, pH 6.2, 50 mM potassium chloride) was performed using Vivaspin centrifugal concentrators (2 kDa molecular weight cut-off). Purity of SL5a was verified by denaturing PAA gel electrophoresis and homogenous folding was monitored by native PAA gel electrophoresis, loading the same RNA concentration as used in NMR experiments.
Using this protocol, two NMR samples of SL5a, an 810 µM uniformly 15N- and a 680 µM uniformly 13C,15N-labeled sample, were prepared and used for the assignment presented herein.
NMR experiments
NMR experiments using the 15N-labeled RNA were carried out at the Karolinska Institute (KI) using a Bruker AVANCEIII 600 MHz NMR spectrometer equipped with a 5 mm, z-axis gradient 1H [13C, 15N, 31P]-QCI cryogenic probe. All NMR experiments with the 13C,15N-labeled RNA were conducted at the Center for Biomolecular Magnetic Resonance (BMRZ) at the Goethe University (GU) Frankfurt using Bruker AVIIIHD NMR spectrometers from 600 to 800 MHz, which are equipped with the following cryogenic probes: 5 mm, z-axis gradient 1H [13C,31P]-TCI cryogenic probe (600 MHz), 5 mm, z-axis gradient 1H [13C, 15N, 31P]-QCI cryogenic probe (700 MHz) and 13C-optimized 5 mm, z-axis gradient 13C, 15N [1H]-TXO cryogenic probe (800 MHz).
At BMRZ and KI, experiments were performed at 298 K if not indicated otherwise. NMR spectra were processed and analyzed using Topspin versions 4.0.8 (GU) and 3.6.2 (KI). The chemical shift assignment was conducted using Sparky (Lee et al. 2015). NMR data were managed and archived using the platform LOGS (2020, version 2.1.54, Signals GmbH & Co KG, www.logs.repository.com). 1H chemical shifts were referenced externally to DSS, and 13C and 15N chemical shifts were indirectly referenced from the 1H chemical shift as described earlier (Wishart et al. 1995).
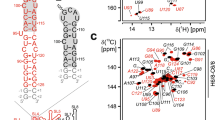
We have previously reported the imino and cytidine amino resonance assignment of SL5a (Wacker et al. 2020) that allowed us to determine the base pairing in this RNA element. The location of stable base pairs is confirmed by through space 2hJNN coupling constants (Dingley et al. 2008) reported in Suppl. Table S1. These assignments were available from experiments conducted on a 15N-labeled RNA sample and provided starting points of the aromatic proton resonance assignment using 1H,1H-NOESY (Tables 1 I, 2 I) and (H)C(CCN)H (Tables 1 IV, 2 V) experiments linking the imino proton resonances to the aromatic protons and carbons (Fig. 1a and b). The remaining H6/8–C6/8 resonances in the aromatic 1H,13C-HSQC spectrum (Tables 1 II, 2 III) were assigned using a 3D 13C-NOESY-HSQC experiment (Table 1 VII), which was selective for the aromatic region. Cytidine and uridine C5-H5 resonances were assigned using 1H,1H-TOCSY (Table 1 VI, Fig. 1e) and 1H,13C-HSQC spectra (Table 1 III, Fig. 1d). Furthermore, quaternary carbon atoms were assigned using an HNCO type experiment (Table 2 IV) and the TROSY relayed HCCH-COSY experiment (Table 1 VIII). The 13C-detected 3D CNC spectrum (Table 1 V, Fig. 1c) linked the aromatic carbons to the anomeric C1′ resonances, where the nitrogen dimension aided in distinguishing between purine and pyrimidine nucleotides as well as between uridines and cytidines. Also, by correlating C6/8 to C1′, resonance overlap is minimized given the broader signal distribution in the carbon as opposed to the respective proton dimensions. Based on C1′ resonances obtained from the CNC spectrum and from sequential assignment in the NOESY spectra, H1′–C1′ correlations were assigned in the 1H,13C-HSQC spectrum (Table 1 III, Fig. 1f). A continuous sequential walk of H1′-to-H6/H8 was possible for both helices (Fig. 1c). The H1′–C1′ assignment was further confirmed with a 3D 13C-NOESY-HSQC experiment (Table 1 IX), which was selective for the C1′ resonances. Using two different 3D HCCH TOCSY experiments (Table 1 X, XI and XII), the remaining ribose carbon resonances C2′–C5′ were assigned. The two experiments differed in the TOCSY mixing time such that with a short mixing time of 6 ms, C2′ and C3′ resonances could be distinguished by intensity differences, while with a long mixing time of 18 ms also C4′ and C5′ carbons were correlated to the C1′ resonances.
Resonance assignment of aromatic protons and carbons and the linkage to the ribose. a HCCNH experiment correlating the imino protons of guanosines and uridines to the corresponding intranucleobase C8 and C6 resonances, respectively. b 1H,13C-HSQC spectrum showing the aromatic H6/8–C6/8 correlations. c 2D plane of the 13C-detected CNC-experiment correlating C6/8 to C1′. d Transposed 1H,13C-HSQC spectrum showing the H5–C5 correlations for uridines and cytidines. e 1H,1H-TOCSY spectrum linking H5 and H6 in pyrimidines. f Transposed 1H,13C-HSQC spectrum of the H1′–C1′ region. Panel c further shows the secondary structure of SL5a with genomic numbering. Positive contours are given in black, negative contours are held in red. Experimental details are given in Table 1. Exemplary connections between the displayed spectra are demonstrated with the gray dashed lines for residues G213 and U191. Assignments of the asymmetric bulge and the apical loop are highlighted with bold font
The U-rich bulge
One of the structural features of the SL5a RNA is an asymmetric U-rich bulge (Fig. 1c). In this likely more dynamic part of the RNA, a near to complete sequential walk (H6/8 to H6/8 or H1′) was possible and thus, all aromatic H6/8–C6/8 correlations were assigned. With the aromatic assignment at hand, the strong imino resonance of a uridine involved in non-canonical base pairing was assigned to residue U194 using the (H)C(CCN)H experiment at 283 K. From observation of this signal, the formation of a base pairing involving U194 and likely either U211 or U212 is suggested. This is further supported by an imino-to-imino NOE contact between U194 and a non-canonical uridine at 273 K. Furthermore, from the U194 carbon chemical shifts in the HNCO experiment, we conclude that the hydrogen bonding interaction is mediated through the C2 carbonyl group (Fürtig et al. 2003; Ohlenschläger et al. 2004). The existence of a GU- wobble base pair involving residues U195 and G210 has not been confirmed, yet. However, broadened imino proton resonances for an additional guanosine and uridine, which are taking part in non-canonical interactions, are observed at low temperature (283 K).
The 5′-UUUCGU-3′ hexaloop
In addition to the U-rich asymmetric bulge (Fig. 1c), SL5a features a 5′-UUUCGU-3′ hexaloop, which also caps the helix of SL5b in the 5′-UTR. Except for residue U205, all aromatic loop assignments were derived from sequential NOE correlations, e.g. H6/8 to H5 or H1′ to H6/8 sequential contacts. Since the central residues of this loop sequence, 5′-UUCG-3′, resemble a highly abundant and well-characterized tetraloop sequence (Cheong et al. 1990; Fürtig et al. 2004; Nozinovic et al. 2010), we asked, whether structural features of this UUCG tetraloop are also found within the 5′-UUUCGU-3′ hexaloop of SL5a. While the characteristic imino proton resonances of the sheared GU base pair in the 5′-UUCG-3′ tetraloop remained elusive in SL5a spectra (e.g. 1H 1D or 1H,15N-HSQC), 1H,13C-HSQC spectra of the ribose region of SL5a and a 14 nt RNA with a 5′-cUUCGg-3′ tetraloop (secondary structure Suppl. Figure 1B) yielded a similar peak pattern (Fig. 2a and b). Here, it is evident that the chemical shifts of the central two nucleotides of the 5′-UUUCGU-3′ hexaloop, U202 and C203, are in good agreement with the respective counterparts in the 5′-cUUCGg-3′ tetraloop. This observation is also reflected in the canonical coordinates (Ebrahimi et al. 2001; Cherepanov et al. 2010), which suggest the ribofuranosyl ring to adopt the C2′-endo conformation for U202 and C203, while the remaining nucleotides (with a complete ribose carbon assignment) adopt the canonical C3′-endo conformation (Fig. 2c). These spectral data suggest a structural similarity between the middle part of the 5′-UUUCGU-3′ hexa- and 5′-cUUCGg-3′ tetraloop. This might not hold true to the same extent for the flanking residues U201 and G204 as characteristic resonances are absent in the 1H,13C-HSQC spectrum of the ribose region (Fig. 2a and b). Thus, the detailed loop architecture remains subject to further structural investigation.
Comparison of 1H,13C-CT-HSQC spectra of the ribose regions of a SL5a and b a 14 nt RNA with 5′-cUUCGg-3′ tetraloop (Fürtig et al. 2004; Nozinovic et al. 2010). Positive contours are given in black, negative contours in red. Experimental details are given in Table 1. The loop sequences are displayed and U202/U7 and C203/C8 resonances are highlighted in bold font. c Canonical coordinates for all residues of SL5a with a complete carbon ribose assignment. For comparison, the canonical coordinates of residues U7 and C8 of a 14 nt RNA with 5′-UUCG-3′ tetraloop are given in red
Assignment and data deposition
The nearly complete resonance assignment of SL5a builds on the imino resonance assignment published earlier (Wacker et al. 2020). Starting from this assignment, all 33 aromatic H6–C6 and H8–C8 correlations were unambiguously assigned. Furthermore, the H2–C2 correlations of the two adenosines present in this RNA as well as all of the H5–C5 correlations of the uridines and cytidines were unambiguously assigned. In addition, the quaternary carbon atoms of the nucleobases in purines (C2: 77%, C4: 69%, C5: 62% and C6: 92%) and pyrimidines (C2: 15% and C4: 15%) were partially assigned. Here, uridine C2 and C4 resonances as well as guanosine C2 and G-1, G188, G198 and G208 C6 resonances were assigned at 283 K. Also, non-protonated tertiary nitrogen atoms of purines (N3: 15% (only adenosines assigned), N7: 100% and N9: 100%) and pyrimidines (N1: 95% and N3: 80% (cytidines)) were successfully assigned to a large extent. Within the ribose moieties, 91% of the H1′ and 91% of the C1′ atoms were assigned. Within the remaining ribose carbon atoms C2′–C5′, 77% were assigned. In summary, we assigned 97% of the 1H (H6/8, H5, H2, H1′) and 92% of the 13C (C6/8, C5(pyr), C1′) atoms, which are considered most important for an in-depth structural characterization. We updated the BMRB deposition with code 50346.
References
Bodenhausen G, Ruben DJ (1980) Natural abundance Nitrogen-15-NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett 69:185–189
Chen S-C, Olsthoorn RCL (2010) Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology 401:29–41. https://doi.org/10.1016/j.virol.2010.02.007
Cheong C, Varani G, Tinoco I (1990) Solution structure of an unusually stable RNA hairpin, 5GGAC(UUCG)GUCC. Nature 346:680–682
Cherepanov AV, Glaubitz C, Schwalbe H (2010) High-resolution studies of uniformly 13C,15N-labeled RNA by solid-state NMR spectroscopy. Angew Chem Int Ed 49:4747–4750. https://doi.org/10.1002/anie.200906885
Dingley AJ, Grzesiek S (1998) Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J Am Chem Soc 7863:714–718
Dingley AJ, Nisius L, Cordier F, Grzesiek S (2008) Direct detection of N–H[...]N hydrogen bonds in biomolecules by NMR spectroscopy. Nat Protoc 3:242–248. https://doi.org/10.1038/nprot.2007.497
Ebrahimi M, Rossi P, Rogers C, Harbison GS (2001) Dependence of 13 C NMR chemical shifts on conformations of RNA nucleosides and nucleotides. J Magn Reson 150:1–9. https://doi.org/10.1006/jmre.2001.2314
Favier A, Brutscher B (2011) Recovering lost magnetization: polarization enhancement in biomolecular NMR. J Biomol NMR 49:9–15. https://doi.org/10.1007/s10858-010-9461-5
Fürtig B, Richter C, Wöhnert J, Schwalbe H (2003) NMR spectroscopy of RNA. ChemBioChem 4:936–962. https://doi.org/10.1002/cbic.200300700
Fürtig B, Richter C, Bermel W, Schwalbe H (2004) New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR 28:69–79. https://doi.org/10.1023/B:JNMR.0000012863.63522.1f
Guan B-J, Wu H-Y, Brian DA (2011) An optimal cis -replication stem-loop IV in the 5′ untranslated region of the mouse coronavirus genome extends 16 nucleotides into open reading frame 1. J Virol 85:5593–5605. https://doi.org/10.1128/JVI.00263-11
Guillerez J, Lopez PJ, Proux F, Launay H, Dreyfus M (2005) A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc Natl Acad Sci USA 102:5958–5963. https://doi.org/10.1073/pnas.0407141102
Hu B, Guo H, Zhou P, Shi ZL (2020) Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. https://doi.org/10.1038/s41579-020-00459-7
Kay LE, Xu G-Y, Singer AU, Muhandiram DR, Forman-Kay JD (1993) A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J Magn Reson Ser B 101:333–337
Kelly JA, Olson AN, Neupane K, Munshi S, Emeterio JS, Pollack L, Woodside MT, Dinman JD (2020) Structural and functional conservation of the programmed −1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J Biol Chem 295:10741–10748. https://doi.org/10.1074/jbc.AC120.013449
Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327. https://doi.org/10.1093/bioinformatics/btu830
Madhugiri R, Fricke M, Marz M, Ziebuhr J (2016) Coronavirus cis-acting RNA elements. Adv Virus Res 96:127–163
Manfredonia I, Nithin C, Ponce-Salvatierra A, Ghosh P, Wirecki TK, Marinus T, Ogando NS, Snijder EJ, Van HMJ, Bujnicki JM, Incarnato D (2020) Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa1053
Martin S, Blankenship C, Rausch JW, Sztuba-Solinska J (2019) Using SHAPE-MaP to probe small molecule-RNA interactions. Methods 167:105–116. https://doi.org/10.1016/j.ymeth.2019.04.009
Masters PS (2019) Coronavirus genomic RNA packaging. Virology 537:198–207
Nozinovic S, Fürtig B, Jonker HRA, Richter C, Schwalbe H (2010) High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res 38:683–694. https://doi.org/10.1093/nar/gkp956
Ohlenschläger O, Wöhnert J, Bucci E, Seitz S, Häfner S, Ramachandran R, Zell R, Görlach M (2004) The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12:237–248. https://doi.org/10.1016/j.str.2004.01.014
Piotto M, Saudek V, Sklen V (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2:661–665
Rangan R, Zheludev IN, Das R (2020) RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses. RNA 26:937–959
Richter C, Kovacs H, Buck J, Wacker A, Fürtig B, Bermel W, Schwalbe H (2010) 13C-direct detected NMR experiments for the sequential J-based resonance assignment of RNA oligonucleotides. J Biomol NMR 47:259–269. https://doi.org/10.1007/s10858-010-9429-5
Schürer H, Lang K, Schuster J, Mörl M (2002) A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res 30:e56
Shaka AJ, Hwang T-L (1995) Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed field gradients. J Magn Reson Ser A 112:275–279
Shaka AJ, Lee CJ, Pines A (1988) Iterative schemes for bilinear operators; application to spin decoupling. J Magn Reson 77:274–293
Simon B, Zanier K, Sattler M (2001) A TROSY relayed HCCH-COSY experiment for correlating adenine H2/H8 resonances in uniformly 13C-labeled RNA molecules. J Biomol NMR 20:173–176
Sklenar V, Peterson RD, Rejante MR, Feigon J (1993a) Two- and three-dimensional HCN experiments for correlating base and sugar resonances in lSN,13C-labeled RNA oligonucleotides. J Biomol NMR 3:721–727
Sklenar V, Piotto M, Leppik R, Saudek V (1993b) Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J Magn Reson Ser A 102:241–245
Sklenár V, Dieckmann T, Butcher SE, Feigon J (1996) Through-bond correlation of imino and aromatic resonances in 13C-, 15N-labeled RNA via heteronuclear TOCSY. J Biomol NMR 7:83–87
Solyom Z, Schwarten M, Geist L, Konrat R, Willbold D, Brutscher B (2013) BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR 55:311–321. https://doi.org/10.1007/s10858-013-9715-0
Tidu A, Janvier A, Schaeffer L, Sosnowski P, Kuhn L, Hammann P, Westhof E, Eriani G, Martin F (2020) The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. https://doi.org/10.1261/rna.078121.120
V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V (2020) Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. https://doi.org/10.1038/s41579-020-00468-6
Vuister W, Bax AD (1992) Resolution enhancement and spectral editing of uniformly 13C-enriched proteins by homonuclear broadband 13C decoupling. J Magn Reson 98:428–435
Wacker A, Weigand JE, Akabayov SR, Altincekic N, Bains K, Banijamali E, Binas O, Castillo-martinez J, Cetiner E, Chiu L, Davila-Calderon J, Dhamotharan K, Duchardt-Ferner E, Ferner J, Frydman L, Fürtig B, Hacker C, Haddad C, Haehnke M, Hengesbach M, Hiller F, Hohmann KF, Hymon D, De JV, Jonker H, Luo L, Mertinkus KR, Keller H, Knezic B, Landgraf T, Löhr F, Muhs C, Novakovic M, Oxenfarth A, Palomino-Schätzlein M, Petzold K, Peter SA, Pyper DJ, Qureshi NS, Riad M, Richter C, Saxena K, Schamber T, Scherf T, Schlagnitweit J, Schlundt A, Schnieders R, Schwalbe H, Simba-lahuasi A, Sreeramulu S, Stirnal E, Sudakov A, Tants J-N, Tolbert BS, Vögele J, Weiß L, Wirmer-Bartoschek J, Wirz Martin MA, Wöhnert J, Zetzsche H (2020) Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa1013
Wishart DS, Bigam CG, Yao J, Abildgaard F, Jane HD, Oldfield E, Markley JL, Sykes BD (1995) Chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140
Wöhnert J, Dingley AJ, Stoldt M, Görlach M, Grzesiek S, Brown LR (1999) Direct identification of NH ··· N hydrogen bonds in non-canonical base pairs of RNA by NMR spectroscopy. Nucleic Acids Res 27:3104–3110
Wöhnert J, Görlach M, Schwalbe H (2003) Triple resonance experiments for the simultaneous correlation of H6/H5 and exchangeable protons of pyrimidine nucleotides in 13C,15N-labeled RNA applicable to larger RNA molecules. J Biomol NMR 26:79–83
Acknowledgements
Work at BMRZ is supported by the state of Hesse. Work in Covid19-nmr was supported by the Goethe Corona Funds, by the IWB-EFRE-programme 20007375 of state of Hesse, the DFG in CRC902: “Molecular Principles of RNA-based regulation.” and infrastructure funds (Project Numbers: 277478796, 277479031, 392682309, 452632086, 70653611). R.S. was recipient of a stipend of the Fonds der Chemischen Industrie. H.S. and B.F. are supported by the DFG in graduate school CLIC (GRK 1986). Work at KI is supported by the Karolinska Institute, Department of Medical Biochemistry and Biophysics for the purchase of a 600-MHz Bruker NMR spectrometer.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schnieders, R., Peter, S.A., Banijamali, E. et al. 1H, 13C and 15N chemical shift assignment of the stem-loop 5a from the 5′-UTR of SARS-CoV-2. Biomol NMR Assign 15, 203–211 (2021). https://doi.org/10.1007/s12104-021-10007-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-021-10007-w