Abstract
Purpose
Metal Regulatory Transcription Factor 1 (MTF1) can be an essential transcription factor for heavy metal response in cells and can also reduce oxidative and hypoxic stresses in cells. However, the current research on MTF1 in gastric cancer is lacking.
Methods
Bioinformatics techniques were used to perform expression analysis, prognostic analysis, enrichment analysis, tumor microenvironment correlation analysis, immunotherapy Immune cell Proportion Score (IPS) correlation and drug sensitivity correlation analysis of MTF1 in gastric cancer. And qRT-PCR was used to verify MTF1 expression in gastric cancer cells and tissues.
Results
MTF1 showed low expression in gastric cancer cells and tissues, and low expression in T3 stage compared with T1 stage. KM prognostic analysis showed that high expression of MTF1 was significantly associated with longer overall survival (OS), FP (first progression) and PPS (post-progression survival) in gastric cancer patients. Cox regression analysis showed that MTF1 was an independent prognostic factor and a protective factor in gastric cancer patients. MTF1 is involved in pathways in cancer, and the high expression of MTF1 is negatively correlated with the half maximal inhibitory concentration (IC50) of common chemotherapeutic drugs.
Conclusion
MTF1 is relatively lowly expressed in gastric cancer. MTF1 is also an independent prognostic factor for gastric cancer patients and is associated with good prognosis. It has the potential to be a diagnostic and prognostic marker for gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is a malignant tumor of the digestive system, which is the second leading cause of cancer death in the world, and has brought a heavy medical burden to the eastern and western people [1, 2]. The 5-year survival rate for patients with gastric cancer is not high after diagnosis, and medical resources are further strained amid the novel coronavirus pandemic [3]. Early gastric cancer can be cured by surgical treatment, and the 5-year survival rate of patients with advanced gastric cancer is less than 10% [4]. Targeted immunotherapy has achieved good feedback in melanoma [5, 6]. The overall response rate (ORR) of melanoma patients treated with nivolumab is 31.7%, which is much higher than that of chemotherapy-only patients with an overall response rate (ORR) of 10.6% [5, 7]. However, due to the lack of more targeted therapeutic targets, targeted therapy in gastric cancer is currently slow progress [8]. It is necessary to dig out potential therapeutic targets and evaluate the risk factors of patients’ prognosis [9].
Metallothionein (MTs) is a low molecular weight protein with high cysteine content, which has a strong affinity for heavy metals [10]. When combined with heavy metals, MTs can regulate the proliferation of normal cells and cancer cells, and participate in tumor formation and progression [11, 12]. Metal Regulatory Transcription Factor 1 (MTF1), as a protein-coding gene, can activate the transcription of Metallothioneins (MTs) gene by binding to Metal response elements (MRE) [13, 14]. At present, the expression of MTF1 in gastric cancer, its effect on the prognosis of gastric cancer patients and its possible role in the development of gastric cancer are not clear. In this study, the expression of MTF1 in gastric cancer cells and tissues of gastric cancer patients was collected and determined. The Kaplan–Meier plotter website was used to collect the relationship between the high and low expression of MTF1 and the prognosis of gastric cancer patients [15]. The correlation between MTF1 and immune cells, immune checkpoint inhibitor genes, and chemotherapeutic drug sensitivity in gastric cancer was analyzed to explore the potential value of MTF1 as a therapeutic target and prognostic marker for gastric cancer.
Materials and methods
Cell culture
Human gastric epithelial cell line (GES-1) and human gastric cancer cell line (MGC-803 and MKN-45) were purchased from the Cell Resource Center of the Chinese Academy of Sciences. All cells were cultured in RPMI 1640 medium (Vivacell, Shanghai, China) containing 10% fetal bovine serum (Biological Industries, Israel).
Ethics and stomach adenocarcinoma (STAD) clinical tissue collection
In August 2022, ten pairs of resected tumor tissues and adjacent tissues of patients undergoing radical gastrectomy were collected in Gansu Provincial Hospital (Lanzhou, Gansu, China). This study was approved by the Medical Ethics Committee of Gansu Provincial Hospital.
qRT-PCR
Under the condition of no enzyme, the total RNA of each cell was extracted using TRIzol reagent (ThermoFisher, Massachusetts, USA) in strict accordance with the instructions for reagent use. RNA extracted using NanoDrop measurement (ThermoFisher, Massachusetts, USA), A260 / A280 between 1.9 and 2.1 RNA is considered to have qualified quality RNA. The purified RNA samples were reverse transcribed into cDNA using reverse transcription kit (vazyme, Nanjing, China), and quantitative real-time polymerase chain reaction (qRT-PCR) was performed using Premixed ROX dye SYBR Green qPCR Master Mix (vazyme, Nanjing, China) on light instrument. GAPDH was used as an internal reference gene, and the gene expression level was calculated using 2− ΔΔCt method. Differences in relative expression levels were analyzed using the t-test, and p < 0.05 was considered statistically significant.
Public data collection and preprocessing
A total of 443 STAD samples were downloaded from the TCGA database (https://portal.gdc.cancer.gov/) and 217 STAD samples (GSE26942) were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/) [16, 17]. The data were normalized using the R software 4.0.1 [18], and differences in MTF1 expression between tumor and cancer tissues were calculated using the Wilcox test, and a value of p < 0.05 was considered statistically significant. The clinical information of the patients corresponding to the samples was collected, and the samples with incomplete clinical information were not included in the correlation analysis. The alteration data of 434 patients with gastric cancer were collected from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/) [19], database to calculate the mutation rate of MTF1 in these samples, and to calculate the statistical difference in the prognostic survival time between the MTF1 altered group and the MTF1 unaltered group, p < 0.05 was considered statistically significant. Data on SATD patients receiving immunotherapy were obtained from The Cancer Imaging Archive (https://www.cancerimagingarchive.net/), and the results showed differences using IPS as an indicator [20].
Correlation of MTF1 expression with prognostic analysis and clinical features in STAD
OS, FP and PPS analysis were performed using the mRNA information of gastric cancer samples in the Kaplan Meier plotter (http://kmplot.com/), and the corresponding curves were plotted. The expression of MTF1 in patients with different age, gender, grade and stage was analyzed by using limma software package and ggpubr software package in R software and the corresponding box plot was drawn [21, 22], and p < 0.05 was considered statistically significant. Univariate Cox regression analysis and multivariate Cox regression analysis were used to explore whether MTF1 was related to the prognosis of patients and to determine whether MTF1 could be used as an independent prognostic factor for STAD.
Multivariate analysis
Group differences were examined with regard to group, age, gender, stage, grade, T, N and M. The descriptive measures were expressed as frequency and proportion for categorical variables, and mean ± standard deviations (SDs) or median and interquartile for continuous variables. The differences between groups were analyzed using the Chi-square test for categorical variables and the t-test (normal distribution) or Kruskal–Wallis rank sum test (non-normal distribution) for continuous variables. An initial model was adjusted only for matching variables. Additionally, a second model was adjusted for gender, age, grade, group, T, N and M. p < 0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using Empower (R) (http://empowerstats.com/en/; X&Y Solutions, Inc., Boston, MA, USA) and R (https://www.R-project.org).
Correlation of MTF1 expression with immune cell infiltration and immune checkpoint genes
The relative content of immune cells in each sample was calculated, and the samples were divided into two groups according to the expression of MTF1. The correlation between MTF1 expression and immune cell infiltration was analyzed by spearman method. Pearson method was used to calculate the correlation between MTF1 and immune checkpoint genes in turn, and the relevant graphs were drawn, and p < 0.05 was considered statistically significant.
Gene set enrichment analysis
All samples were divided into high and low expression groups according to the expression of MTF1 in the samples. The c5.go.v7.4.symbols.gmt file and c2.cp.kegg.v7.4.symbols.gmt file were downloaded from the Molecular Signatures Database [23], and The Gene Ontology (GO) enrichment analysis and Genes and Genomes (KEGG) enrichment analysis were performed to elucidate the biological processes and signaling pathways that MTF1 may act on [24,25,26].
Sensitivity analysis of chemotherapeutic drugs
The data of drug sensitivity of tumor cells were obtained from the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/), and the IC50 and mRNA expression information of various compounds acting on tumor cells were obtained [27]. The Pearson method was used to correlate IC50 values with mRNA expression to show the correlation between MTF1 and sensitivity to common chemotherapy drugs [28].
Statistics
Wilcoxon test was performed on the expression of MTF1 in gastric cancer tissues and adjacent tissues obtained from GEO database. The t test was performed on the MTF1 expression data conforming to the normal distribution and the homogeneity of variance. The t ‘test was performed on the MTF1 expression data conforming to the normal distribution and the heterogeneity of variance. The non-parametric test was performed on the data that did not conform to the normal distribution. Analysis of Variance (ANOVA) was used to calculate the expression of MTF1 in different cells. The limma software package was used to calculate the correlation between the expression of MTF1 and the clinical characteristics of gastric cancer patients. Spearman Correlation Analysis was used to calculate the correlation between MTF1 expression and immune cell infiltration. Pearson Correlation Analysis was used to calculate the correlation between MTF1 expression and immune checkpoint inhibitor gene expression and the correlation with chemotherapy drug sensitivity.
Results
MTF1 is relatively low in STAD
By analyzing the differences of samples in GEO database, we found that MTF1 was relatively low expressed in gastric cancer tissues compared with normal tissues (Fig. 1A). The mRNA expression of MTF1 in gastric cancer cells and gastric epithelial cells was detected by qRT-PCR. Although there was no difference in the mRNA expression level of MTF1 between MKN45 and GES1, the expression of MTF1 in MGC803 was significantly lower than that in GES-1 at the mRNA level (p < 0.001) (Fig. 1B). The results of measuring the mRNA expression level of MTF1 in gastric cancer tissues also showed that MTF1 was relatively low in gastric cancer tissues compared to adjacent tissues (Fig. 1C).
The expression of MTF1 in gastric cancer tissues and adjacent tissues in GSE26942 was analyzed by Wilcoxon test in R 4.0.3 software A ANOVA was used to analyze the expression of MTF1 in gastric cancer cells and gastric epithelial cells in GraphPad Prism 8.0.2 software B Mann–Whitney test was used to analyze the expression of MTF1 in gastric cancer tissues and adjacent tissues in GraphPad Prism 8.0.2 software (C). The frequency of alteration in MTF1 (D) and the OS-based KM prognostic analysis of the MTF1 alterated group and the unalterated group were obtained on the cBioPortal website (E). OS overall survival, KM Kaplan–Meier
MTF1 alteration in STAD samples
Among the 434 patients with gastric cancer, 3.69% of the samples had MTF1 gene mutation (Fig. 1D). The main mutation type of MTF1 in STAD samples was Mutation (2.07%), followed by Amplification (1.38%), and the smallest mutation type was Deep Deletion (0.23%). KM prognostic analysis of patients (Fig. 1E) with MTF1 alter group and MTF1 unatler group showed that patients with MTF1 mutation group had longer OS (Tables 1, 2 and 3).
MTF1 expression is associated with T stage in STAD patients
To explore the effect of MTF1 expression level on the clinical characteristics of STAD patients (Fig. 2A), we analyzed the expression of MTF1 in different ages (Fig. 2B), different gender (Fig. 2C), different grades (Fig. 2D) and different TNM stages (Fig. 2E–H). It was found that MTF1 was highly expressed in T1, especially compared to T3 (Fig. 2F). Multivariate analysis showed that MTF1 expression significantly affected the stage of gastric cancer patients (Tables 4 and 5).
The expression of MTF1 in different clinical feature subgroups (A), different age groups (B), different genders (C), different Grades (D), different stages (E), different T staging (F), different M staging (G) and different N staging (H) were analyzed by 'limma' package. T tumor, N lymph node; M metastasis
High MTF1 expression correlates with good prognosis in STAD patients
According to the results of Kaplan–Meier survival analysis, the high expression of MTF1 was significantly correlated with the better prognosis of patients. According to the median value of expression, STAD patients were divided into high expression MTF1 group and low expression MTF1 group. The high expression of MTF1 was significantly correlated with longer OS in STAD patients (p = 0.0016, Fig. 3A), longer FP in SATD patients (p = 0.0042, Fig. 3B) and longer PPS in SATD patients (p = 0.0025, Fig. 3C). Multivariate cox regression analysis and univariate cox regression analysis also verified that MTF1 expression can be used as an independent prognostic factor for OS in SATD patients. Multivariate analysis showed that the high expression of MTF1 was significantly correlated with the longer OS of STAD patients (p = 0.018, Fig. 3D). Univariate analysis also showed that the high expression of MTF1 was significantly correlated with the longer OS of STAD patients (p = 0.043, Fig. 3E).
KM prognostic analysis of the correlation between MTF1 expression and OS (A), FP (B), and PPS (C) in gastric cancer patients was obtained on the Kaplan–Meier Plotter website. Multivariate Cox regression analysis (D) and univariate Cox regression analysis (E) of MTF1 as an independent prognostic factor for gastric cancer patients were tested using the ‘survival’ software package. KM Kaplan–Meier, OS overall survival, FP first progression, PPS post-progression survival
MTF1 is involved in biological processes and signaling pathways in STAD
MTF1 is mainly involved in Gene Ontology term by epidermis development, skin development and keratinocyte differentiation (Fig. 4A). The main signaling pathway involved in MTF1 is epidermis development, skin development and regulation of body fluid levels (Fig. 4B). GSEA results showed that the main active biological processes in MTF1 high expression group were cornification, epidermal cell differentiation, epidermis development and keratinization, and the main active biological processes in MTF1 low expression group were detection of chemical stimulus (Fig. 4C). The main active signaling pathway in MTF1 high expression group is pathways in cancer, and the main active signaling pathway in MTF1 low expression group is complement and coagulation cascades, olfactory transduction, oxidative phosphorylation and ribosome (Fig. 4D).
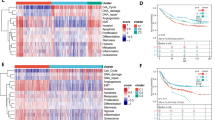
MTF1 is associated with immune cell infiltration
Correlation analysis between MTF1 and immune cell infiltration (Fig. 5A) showed that MTF1 expression was significantly positively correlated with Dendritic cells activated (Fig. 5B), NK cells resting (Fig. 5C), Neutrophils (Fig. 5D) and T cells CD4 memory resting (Fig. 5E). There was a significant negative correlation with B cells naive (Fig. 5F), T cells CD8 (Fig. 5G) and NK cells activated (Fig. 5H).
Spearman Correlation Analysis was used to analyze the correlation between MTF1 expression and immune cell infiltration in R 4.0.3 software (A). Spearman Correlation Analysis was used to analyze the correlation between MTF1 expression and Dendritic cells activated (B), NK cells resting (C), Neutrophils (D), T cells CD4 memory resting (E), B cells naive (F), T cells CD8 (G) and NK cells activated (H). NK nature killer
MTF1 is associated with immune checkpoint inhibitor gene expression and immunotherapy
According to the results of spearman statistical method, the expression of MTF1 was significantly correlated with the expression of 16 immune checkpoint inhibitor genes, and generally positively correlated (Fig. 6A). From the difference of IPS between the two groups with high and low expression of MTF1, although there is no significant difference between the expression of MTF1 and IPS in CTLA4 (−) _ PD1 ( +) group (Fig. 6B) and CTLA4 ( +) _ PD1 ( +) group (Fig. 6D), the low expression group of MTF1 had higher IPS in CTLA4 ( +) _ PD1 (−) group (Fig. 6C) and CTLA4 (−) _ PD1 (−) group (Fig. 6E).
MTF1 is associated with chemosensitivity
According to the pRRophetic algorithm analysis results, the IC50 of nine chemotherapeutic drugs was significantly positively correlated with the expression of MTF1. The IC50 of 50 chemotherapeutic drugs was significantly negatively correlated with the expression of MTF1. See Annex 1 for details.
Discussion
We verified that MTF1 was relatively low in gastric cancer cells and gastric cancer tissues firstly, compared with normal gastric epithelial cells and adjacent tissues. Although there are individual differences in the expression of MTF1 in surgically resected gastric cancer tissues, in general, it is relatively low in gastric cancer tissues, which is consistent with the expression trend of MTF1 in surgically resected lung cancer tissues and uterine endometrial cancer cells [29, 30]. MTF1 expression was relatively low in gastric cancer tissues, gastric cancer cell MGC803, and gastric cancer tissues from the geo database, suggesting that MTF1 has the potential to as a diagnostic biomarker for gastric cancer. The results of KM prognostic analysis (HR = 0.68), univariate Cox regression analysis (HR = 0.568) and multivariate Cox regression analysis (HR = 0.616) showed that MTF1 was a protective factor for gastric cancer patients, and the high expression of MTF1 was a good prognostic marker for gastric cancer patients. We also found that the expression of MTF1 in T1 stage was significantly higher than that in T3 stage, suggesting that MTF1 may inhibit tumor progression. Multivariate analysis results also support that MTF1 expression affects the stage of gastric cancer. Analysis in the cBioPortal database showed that gastric cancer patients with MTF1 mutation had a relatively longer OS, and the genetic alterations of MTF1 in gastric cancer samples were mainly mutation and amplification. The mutation and amplification of MTF1 may increase the transcription of MTF1, and the prolongation of OS in patients with gastric cancer may also be due to the increased transcription of MTF1, a protective factor. The results of drug sensitivity analysis showed that MTF1 expression tended to be associated with increased drug sensitivity (in 50 drugs) rather than decreased drug sensitivity (in 9 drugs). Higher drug sensitivity may be the reason why OS and FP in patients with high MTF1 expression were longer than those in patients with low MTF1 expression.
The infiltration of seven immune cells was significantly correlated with the expression of MTF1, including Dendritic cells activated, NK cells resting, Neutrophils, T cells CD4 memory resting, B cells navie, T cells CD8 and NK cells activated. In particular, dendritic cells activated, NK cells resting, Neutrophils and T cells CD4 memory resting were significantly positively correlated with MTF1 expression, suggesting that MTF1 has an effect on tumor immunity in gastric cancer. GESA based on KEGG showed that the differential genes of the two groups with high and low expression of MTF1 were not only directly involved in pathways in cancer (under the classification criteria of KEGG), but also involved in oxidative phosphorylation and ribosome, which was consistent with the previous view that MTF1 could make cells less subject to oxidative and hypoxic stresses [31, 32]. The results showed that the expression of MTF1 was significantly positively correlated with the expression of 16 immune checkpoint inhibitor gene, and was significantly negatively correlated with the IPS of immunotherapy in CTLA4 ( +) _PD1 (−) group and CTLA4 (−) _PD1 (−) group. The high expression of MTF1 may have a negative effect on gastric cancer tumor immunity, but the mechanism of MFT1 affecting tumor immunity and immunotherapy is not clear. Another shortcoming of this study is that the included samples may not be sufficient. The expression of MTF1 in patients has certain individual differences, and only the expression of MTF1 at the mRNA level is analyzed, and the expression of MTF1 at the protein level is not verified.
In summary, we identified for the first time that the expression of MTF1 in gastric cancer cells MGC803 was significantly lower than that in normal gastric epithelial cells, and verified the relatively low expression of MTF1 in gastric cancer tissues in 10 pairs of clinical tissues, and included 217 gastric cancer patients in the GEO database. The expression of MTF1 in different clinical feature subgroups was analyzed, and the low expression of MTF1 in T3 stage was found. More importantly, KM prognostic analysis and Cox regression analysis showed that MTF1 was an independent prognostic factor for gastric cancer patients and was likely to be a protective factor. The enrichment analysis, tumor immune infiltration and drug sensitivity analysis provided clues for the possible biological function and possible mechanism of MTF1 in gastric cancer.
Conclusion
The mRNA level of MTF1 was relatively low in gastric cancer cells and gastric cancer tissues, as well as in T3 staging, and the high expression of MTF1 was significantly correlated with longer OS, FP and PPS in gastric cancer patients, as well as the increased drug sensitivity of 50 common chemotherapeutic drugs. MTF1 has the potential as a diagnostic and prognostic marker for gastric cancer.
Data availability
The datasets analyzed during the current study are available in GEO database (https://www.ncbi.nlm.nih.gov/), TCGA database (https://portal.gdc.cancer.gov/), The Cancer Imaging Archive (https://www.cancerimagingarchive.net/), Kaplan Meier plotter portal (https://kmplot.com/analysis/), cBioPortal database (http://cbioportal.org), The Molecular Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb) and GDSC database(https://www.cancerrxgene.org/).
References
Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71(3):264–79.
Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;18(3):534–42.
Kawazoe A, Shitara K, Boku N, Yoshikawa T, Terashima M. Current status of immunotherapy for advanced gastric cancer. Jpn J Clin Oncol. 2021;51(1):20–7.
Li GZ, Doherty GM, Wang J. Surgical management of gastric cancer: a review. JAMA Surg. 2022;157(5):446–54.
Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445(7130):851–7.
Sullivan RJ. The role of targeted therapy for melanoma in the immunotherapy era. Semin Cutan Med Surg. 2018;37(2):112–9.
Ji L, Zhao G, Zhang P, Huo W, Dong P, Watari H, et al. Knockout of MTF1 inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer. 2018;9(24):4578–85.
Kole C, Charalampakis N, Tsakatikas S, Kouris NI, Papaxoinis G, Karamouzis MV, et al. Immunotherapy for gastric cancer: a 2021 update. Immunotherapy. 2022;14(1):41–64.
Matsuzaki J, Tsugawa H, Suzuki H. Precision medicine approaches to prevent gastric cancer. Gut Liver. 2021;15(1):3–12.
Aalami AH, Hoseinzadeh M, Hosseini Manesh P, Jiryai Sharahi A, Kargar AE. Carcinogenic effects of heavy metals by inducing dysregulation of microRNAs: a review. Mol Biol Rep. 2022;49(12):12227–38.
Si M, Lang J. The roles of metallothioneins in carcinogenesis. J Hematol Oncol. 2018;11(1):107.
Helal NS, Moussa MM, Aboushousha T, Aboul-Ezz MA, Magdy M. Differential expression of metallothionein and p21 in gastric cancer and some precursor lesions. Eur Rev Med Pharmacol Sci. 2022;26(9):3100–8.
Dong G, Chen H, Qi M, Dou Y, Wang Q. Balance between metallothionein and metal response element binding transcription factor 1 is mediated by zinc ions (review). Mol Med Rep. 2015;11(3):1582–6.
Westin G, Schaffner W. A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. EMBO J. 1988;7(12):3763–70.
Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and Implementation. J Med Internet Res. 2021;23(7): e27633.
Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19(1A):A68-77.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–5.
The R Development Core Team. R: A language and environment for statistical computing. Version 2.15.0.2013
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–57.
Kassambara A, Kassambara M. Package ‘ggpubr’, 6. 2020
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7): e47.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40.
Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue):D258–61.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50.
Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–61.
Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9): e107468.
Chen Y. Identification and validation of cuproptosis-related prognostic signature and associated regulatory axis in uterine corpus endometrial carcinoma. Front Genet. 2022;13: 912037.
Liang GY, Lu SX, Xu G, Liu XD, Li J, Zhang DS. Expression of metallothionein and Nrf2 pathway genes in lung cancer and cancer-surrounding tissues. World J Surg Oncol. 2013;11:199.
Grzywacz A, Gdula-Argasińska J, Muszyńska B, Tyszka-Czochara M, Librowski T, Opoka W. Metal responsive transcription factor 1 (MTF-1) regulates zinc dependent cellular processes at the molecular level. Acta Biochim Pol. 2015;62(3):491–8.
Sheng M, Zhang G, Wang J, Yang Q, Zhao H, Cheng X, et al. Remifentanil induces cardio protection against ischemia/reperfusion injury by inhibiting endoplasmic reticulum stress through the maintenance of zinc homeostasis. Anesth Analg. 2018;127(1):267–76.
Acknowledgements
This work has benefited from aforementioned databases. The authors thank the Key Laboratory of Molecular Diagnostics and Precision Medicine for Surgical Oncology in Gansu Province and the DaVinci Surgery System Database (DSSD, www.davincisurgerydatabase.com) for their help and support in the methodology.
Funding
This work was funded by the 2021 Central-Guided Local Science and Technology Development Fund (ZYYDDFFZZJ-1), The 2022 Master/Doctor/Postdoctoral program of NHC Key Laboratory of Diagnosis and Therapy of Gastrointestinal Tumor (NHCDP2022014), Natural Science Foundation of Gansu Province, China (No.18JR3RA052), Lanzhou Talent Innovation and Entrepreneurship Project Task Contract (No. 2016-RC-56), Gansu Da Vinci robot high-end diagnosis and treatment team construction project, and National Key Research and Development Program (No. 2018YFC1311500).
Author information
Authors and Affiliations
Contributions
JH and XJ conceived and designed the study, and drafted the manuscript. MY, PW and LF collected, analyzed and interpreted the experimental data. GZ and HC revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by Ethical Committee of Gansu Provincial Hospital and conducted in accordance with the ethical standards.
Informed consent
Subjects signed the informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12094_2023_3198_MOESM1_ESM.jpg
Supplementary file1 Figure S1: Correlation analysis between MTF1 expression and sensitivity of common chemotherapeutic drugs (JPG 139 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, J., Jiang, X., Yu, M. et al. MTF1 has the potential as a diagnostic and prognostic marker for gastric cancer and is associated with good prognosis. Clin Transl Oncol 25, 3241–3251 (2023). https://doi.org/10.1007/s12094-023-03198-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03198-2