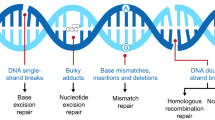
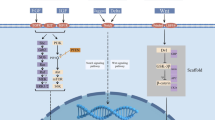
Abstract
Triple-negative breast cancer (TNBC) is the most invasive molecular subtype of breast cancer (BC), accounting for about nearly 15% of all BC cases reported annually. The absence of the three major BC hormone receptors, Estrogen (ER), Progesterone (PR), and Human Epidermal Growth Factor 2 (HER2) receptor, accounts for the characteristic “Triple negative” phraseology. The absence of these marked receptors makes this cancer insensitive to classical endocrine therapeutic approaches. Hence, the available treatment options remain solemnly limited to only conventional realms of chemotherapy and radiation therapy. Moreover, these therapeutic regimes are often accompanied by numerous treatment side-effects that account for early distant metastasis, relapse, and shorter overall survival in TNBC patients. The rigorous ongoing research in the field of clinical oncology has identified certain gene-based selective tumor-targeting susceptibilities, which are known to account for the molecular fallacies and mutation-based genetic alterations that develop the progression of TNBC. One such promising approach is synthetic lethality, which identifies novel drug targets of cancer, from undruggable oncogenes or tumor-suppressor genes, which cannot be otherwise clasped by the conventional approaches of mutational analysis. Herein, a holistic scientific review is presented, to undermine the mechanisms of synthetic lethal (SL) interactions in TNBC, the epigenetic crosstalks encountered, the role of Poly (ADP-ribose) polymerase inhibitors (PARPi) in inducing SL interactions, and the limitations faced by the lethal interactors. Thus, the future predicament of synthetic lethal interactions in the advancement of modern translational TNBC research is assessed with specific emphasis on patient-specific personalized medicine.




Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- ACS:
-
American cancer society
- BC:
-
Breast cancer
- BER:
-
Base excision repair
- CDK:
-
Cyclin-dependent kinase
- ChIP:
-
Chromatin immunoprecipitation sequencing
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- CSC:
-
Cancer stem cells
- DDFS:
-
Distant disease-free survival
- DFS:
-
Disease-free survival
- DDR:
-
DNA damage response
- DSB:
-
Double-strand break
- DSC:
-
Double-strand crosslink
- EMA:
-
European medicines agency
- EMT:
-
Epithelial–mesenchymal transformation
- ER:
-
Estrogen receptor
- gPALB2:
-
Germline PALB2
- gRNA:
-
Guide RNA
- HER2:
-
Human epidermal growth factor receptor 2
- HMG:
-
Mobility group (protein)
- HR:
-
Homologous recombination
- HRR:
-
Homologous recombination repair
- IDFS:
-
Invasive disease-free survival
- lncRNA:
-
Long non-coding RNA
- LOH:
-
Loss of heterozygosity
- MDR:
-
Multidrug resistance
- MHEJ:
-
Microhomology-mediated end joining
- MRD:
-
Minimal residual disease
- NGS:
-
Next-generation sequencing
- NHEJ:
-
Non-homologous end joining
- NLS:
-
Nuclear localizing sequence
- OS:
-
Overall survival
- ORR:
-
Overall response rate
- PARPi:
-
Poly (ADP-ribose) polymerase inhibitor
- PFS:
-
Progression-free survival
- pi-RNA:
-
Piwi-interacting RNA
- PR:
-
Progesterone receptor
- RNAi:
-
RNA interference
- SDL:
-
Synthetic dosage lethality
- SR:
-
Serine and arginine-rich (proteins)
- sgRNA:
-
Single-guide RNA
- shRNA:
-
Short/small hairpin RNA
- si-RNA:
-
Small interfering RNA
- SL:
-
Synthetic lethal
- sBRCA:
-
Somatic BRCA
- SSB:
-
Single-strand break
- TME:
-
Tumor microenvironment
- TNBC:
-
Triple-negative breast cancer
- TSG:
-
Tumor suppressor gene
- WHO:
-
World Health Organization
References
Han Ye, Xiaopeng Yu, Li S, Tian Ye, Liu C. New perspectives for resistance to PARP inhibitors in triple-negative breast cancer. Front Oncol. 2020;10:578095.
Ahmad A, editor. Breast cancer metastasis and drug resistance: challenges and progress. vol. 1152. Springer Nature; 2019.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33.
Dahlin JL, Walters MA. The essential roles of chemistry in high-throughput screening triage. Future Med Chem. 2014;6(11):1265–90.
do Nascimento RG, Otoni KM. Histological and molecular classification of breast cancer: what do we know. Mastology. 2020;30:e20200024.
Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010;32(1–2):35.
Tsang JYS, Huang Y-H, Luo M-H, Ni Y-B, Chan S-K, Lui PCW, Yu AMC, Tan PH, Tse GM. Cancer stem cell markers are associated with adverse biomarker profiles and molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;136:407–17.
Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009;9:S73–81.
Borri F, Granaglia A. Pathology of triple negative breast cancer. Semin Cancer Biol. 2021;72:136–45.
Yin Li, Duan J-J, Bian X-W, Shi-cang Yu. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22:1–13.
Barchiesi G, Roberto M, Verrico M, Vici P, Tomao S, Tomao F. Emerging role of PARP inhibitors in metastatic triple negative breast cancer. Current scenario and future perspectives. Front Oncol. 2021;11:769280.
Nwagu GC, Bhattarai S, Swahn M, Ahmed S, Aneja R. Prevalence and mortality of triple-negative breast cancer in West Africa: biologic and sociocultural factors. JCO Global Oncol. 2021;7:1129–40.
Li S, Topatana W, Juengpanich S, Cao J, Jiahao Hu, Zhang B, Ma D, Cai X, Chen M. Development of synthetic lethality in cancer: molecular and cellular classification. Signal Transduct Target Ther. 2020;5(1):241.
Li C-H, Hsu T-I, Chang Y-C, Chan M-H, Lu P-J, Hsiao M. Stationed or relocating: The seesawing emt/met determinants from embryonic development to cancer metastasis. Biomedicines. 2021;9(9):1265.
Dobzhansky TH. Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics. 1946;31(3):269.
Zecchini V, Frezza C. Metabolic synthetic lethality in cancer therapy. Biochim Biophys Acta (BBA) Bioenerg. 2017;188(8):723–31.
Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349(6255):1483–9.
Zhang B, Tang C, Yao Y, Chen X, Zhou C, Wei Z, Xing F, et al. The tumor therapy landscape of synthetic lethality. Nat Commun. 2021;12(1):1275.
Sajesh BV, Guppy BJ, McManus KJ. Synthetic genetic targeting of genome instability in cancer. Cancers. 2013;5(3):739–61.
de la Cruz FF, Gapp BV, Nijman SMB. Synthetic lethal vulnerabilities of cancer. Annu Rev Pharmacol Toxicol. 2015;55:513–31.
Dwane L, Behan FM, Gonçalves E, Lightfoot H, Yang W, van der Meer D, Shepherd R, Pignatelli M, Iorio F, Garnett MJ. Project Score database: a resource for investigating cancer cell dependencies and prioritizing therapeutic targets. Nucleic Acids Res. 2021;49(D1):D1365–72.
Ma M, Ye AY, Zheng W, Kong L. A guide RNA sequence design platform for the CRISPR/Cas9 system for model organism genomes. BioMed Res Int. 2013;2013:270805. https://doi.org/10.1155/2013/270805
Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21.
Walsh CS. Two decades beyond BRCA1/2: Homologous recombination, hereditary cancer risk and a target for ovarian cancer therapy. Gynecol Oncol. 2015;137(2):343–50.
Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, Livingston DM, Parvin JD. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci. 1997;94(11):5605–10.
Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316(5828):1194–8.
Dhawan M, Ryan CJ. BRCAness and prostate cancer: Diagnostic and therapeutic considerations. Prostate Cancer Prostatic Dis. 2018;21(4):488–98.
Turner N, Tutt A, Ashworth A. Hallmarks of’BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–9.
Liu L, Matsunaga Y, Tsurutani J, Akashi-Tanaka S, Masuda H, Ide Y, Hashimoto R, et al. BRCAness as a prognostic indicator in patients with early breast cancer. Sci Rep. 2020;10(1):1–8.
Tung N, Lin NU, Kidd J, Allen BA, Singh N, Wenstrup RJ, Hartman A-R, Winer EP, Garber JE. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34(13):1460.
Winter C, Nilsson MP, Olsson E, George AM, Chen Y, Kvist A, Törngren T, et al. Targeted sequencing of BRCA1 and BRCA2 across a large unselected breast cancer cohort suggests that one-third of mutations are somatic. Ann Oncol. 2016;27(8):1532–8.
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips K-A, Mooij TM, Roos-Blom M-J, Jervis S, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402–16.
Toss A, Molinaro E, Venturelli M, Domati F, Marcheselli L, Piana S, Barbieri E, et al. BRCA detection rate in an Italian cohort of luminal early-onset and triple-negative breast cancer patients without family history: when biology overcomes genealogy. Cancers. 2020;12(5):1252.
Guo M, Ren J, House MG, Qi Yu, Brock MV, Herman JG. Accumulation of promoter methylation suggests epigenetic progression in squamous cell carcinoma of the esophagus. Clin Cancer Res. 2006;12(15):4515–22.
McLornan DP, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med. 2014;371(18):1725–35.
Yan W, Herman JG, Guo M. Epigenome-based personalized medicine in human cancer. Epigenomics. 2016;8(1):119–33.
Gao A, Guo M. Epigenetic based synthetic lethal strategies in human cancers. Biomark Res. 2020;8(1):1–14.
Gao D, Herman JG, Guo M. The clinical value of aberrant epigenetic changes of DNA damage repair genes in human cancer. Oncotarget. 2016;7(24):37331.
Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92(7):564–9.
Aguilar-Quesada R, Muñoz-Gámez JA, Martín-Oliva D, Peralta A, Valenzuela MT, Matínez-Romero R, Quiles-Pérez R, et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol. 2007;8(1):1–8.
Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10(4):293–301.
Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–74.
Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297(5581):547–51.
Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–35.
Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–45.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–8.
Ashworth A. A synthetic lethal therapeutic approach: poly (ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26(22):3785–90.
Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10(8):1192–9.
Murcia De, Menissier J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Manuel Mark F, Oliver J, et al. Requirement of poly (ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci. 1997;94(14):7303–7.
Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair. 2008;7(7):1077–86.
Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly (ADP-ribose) glycohydrolase in the control of poly (ADP-ribose) metabolism. Exp Cell Res. 2001;268(1):7–13.
Nguewa PA, Fuertes MA, Valladares B, Alonso C, Pérez JM. Poly (ADP-ribose) polymerases: homology, structural domains and functions. Novel therapeutical applications. Prog Biophys Mol Biol. 2005;88(1):143–72.
Virág L, Szabó C. The therapeutic potential of poly (ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54(3):375–429.
Jagtap P, Szabó C. Poly (ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4(5):421–40.
Pillai JB, Russell HM, Raman J, Jeevanandam V, Gupta MP. Increased expression of poly (ADP-ribose) polymerase-1 contributes to caspase-independent myocyte cell death during heart failure. Am J Physiol Heart Circ Physiol. 2005;288(2):H486–96.
Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair. 2007;6(6):695–711.
Amé J-C, Spenlehauer C, De Murcia G. The PARP superfamily. BioEssays. 2004;26(8):882–93.
Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31(6):446–54.
Huber A, Bai P, Murcia JMD, De Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair. 2004;3(8–9):1103–8.
Hoeijmakers JHJ. DNA damage, aging, and cancer. N Engl J Med. 2009;361(15):1475–85.
Haince J-F, McDonald D, Rodrigue A, Déry U, Masson J-Y, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283(2):1197–208.
Gogola E, Duarte AA, de Ruiter JR, Wiegant WW, Schmid JA, de Bruijn R, James DI, et al. Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell. 2018;33(6):1078–93.
Comen EA, Robson M. Poly (ADP-ribose) polymerase inhibitors in triple-negative breast cancer. Cancer J (Sudbury, Mass). 2010;16(1):48.
Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20(3):294–302.
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly (ADP-ribose) polymerase. Nature. 2005;434(7035):913–7.
Rakha EA, El-Sheikh SE, Kandil MA, El-Sayed ME, Green AR, Ellis IO. Expression of BRCA1 protein in breast cancer and its prognostic significance. Hum Pathol. 2008;39(6):857–65.
Wilson CA, Ramos L, Villaseñor MR, Anders KH, Press MF, Clarke K, Karlan B, et al. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet. 1999;21(2):236–40.
Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25(43):5846–53.
Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, Savage K, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26(14):2126–32.
Catteau A, Harris WH, Chun-Fang Xu, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18(11):1957–65.
Rice JuddC, Ozcelik H, Maxeiner P, Andrulis I, Futscher BW. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis. 2000;21(9):1761–5.
Baldassarre G, Battista S, Belletti B, Thakur S, Pentimalli F, Trapasso F, Fedele M, Pierantoni G, Croce CM, Fusco A. Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma. Mol Cell Biol. 2003;23(7):2225–38.
Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, Lidereau R, Mikaélian I, Mazoyer S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3(5):279–90.
Matamala N, Vargas MT, González-Cámpora R, Arias JI, Menéndez P, Andrés-León E, Yanowsky K, et al. MicroRNA deregulation in triple negative breast cancer reveals a role of miR-498 in regulating BRCA1 expression. Oncotarget. 2016;7(15):20068.
Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, Sledge GW, Carey LA. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14(24):8010–8.
Calabrese CR, Almassy R, Barton S, Batey MA, Hilary Calvert A, Canan-Koch S, Durkacz BW, et al. Anticancer chemosensitization and radiosensitization by the novel poly (ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96(1):56–67.
Tentori L, Portarena I, Bonmassar E, Graziani G. Combined effects of adenovirus-mediated wild-type p53 transduction, temozolomide and poly (ADP-ribose) polymerase inhibitor in mismatch repair deficient and non-proliferating tumor cells. Cell Death Differ. 2001;8(5):457–69.
Tentori L, Portarena I, Vernole P, De Fabritiis P, Madaio R, Balduzzi A, Roy R, et al. Effects of single or split exposure of leukemic cells to temozolomide, combined with poly (ADP-ribose) polymerase inhibitors on cell growth, chromosomal aberrations and base excision repair components. Cancer Chemother Pharmacol. 2001;47(4):361–9.
Delaney CA, Wang L-Z, Kyle S, White AW, Calvert AH, Curtin NJ, Durkacz BW, Hostomsky Z, Newell DR. "Potentiation of temozolomide and topotecan growth inhibition and cytotoxicity by novel poly (adenosine diphosphoribose) polymerase inhibitors in a panel of human tumor cell lines. Clin Cancer Res. 2000;6(7):2860–7.
Yung TMC, Sato S, Satoh MS. Poly (ADP-ribosyl) ation as a DNA damage-induced post-translational modification regulating poly (ADP-ribose) polymerase-1-topoisomerase I interaction. J Biol Chem. 2004;279(38):39686–96.
Malanga M, Althaus FR. Poly (ADP-ribose) reactivates stalled DNA topoisomerase I and induces DNA strand break resealing. J Biol Chem. 2004;279(7):5244–8.
O’Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364(3):205–14.
Litton JK, Rugo HS, Ettl J, Hurvitz SA, Gonçalves A, Lee K-H, Fehrenbacher L, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753–63.
Robson ME, Tung N, Conte P, Im S-A, Senkus E, Xu B, Masuda N, et al. OlympiAD final overall survival and tolerability results: Olaparib versus chemotherapy treatment of physician’s choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann Oncol. 2019;30(4):558–66.
Javle M, Curtin NJ. The potential for poly (ADP-ribose) polymerase inhibitors in cancer therapy. Ther Adv Med Oncol. 2011;3(6):257–67.
Murai J, Huang S-Y, Renaud A, Zhang Y, Ji J, Takeda S, Morris J, Teicher B, Doroshow JH, Pommier Y. Stereospecific PARP Trapping by BMN 673 and comparison with Olaparib and RucaparibPARP trapping by BMN 673. Mol Cancer Ther. 2014;13(2):433–43.
Topatana W, Juengpanich S, Li S, Cao J, Jiahao Hu, Lee J, Suliyanto K, et al. Advances in synthetic lethality for cancer therapy: Cellular mechanism and clinical translation. J Hematol Oncol. 2020;13(1):1–22.
Beijersbergen RL, Wessels LFA, Bernards R. Synthetic lethality in cancer therapeutics. Annu Rev Cancer Biol. 2017;1:141–61.
Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–79.
Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AOH, Zander SAL, Derksen PWB, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci. 2008;105(44):17079–84.
Willers H, Taghian AG, Luo C-M, Treszezamsky A, Sgroi DC, Powell SN. Utility of DNA repair protein foci for the detection of putative BRCA1 pathway defects in breast cancer biopsies. Mol Cancer Res. 2009;7(8):1304–9.
Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451(7182):1111–5.
Yazinski SA, Comaills V, Buisson R, Genois M-M, Nguyen HD, Ho CK, Kwan TT, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31(3):318–32.
Coussy F, El-Botty R, Château-Joubert S, Dahmani A, Montaudon E, Leboucher S, Morisset L, et al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci Transl Med. 2020;12(531):eaax2625.
Zimmer AS, Gillard M, Lipkowitz S, Lee J-M. Update on PARP inhibitors in breast cancer. Curr Treat Options Oncol. 2018;19:1–19.
Tung NM, Robson ME, Ventz S, Santa-Maria CA, Nanda R, Marcom PK, Shah PD, et al. TBCRC 048: phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol. 2020;38(36):4274–82.
Li Y, Zhan Z, Yin X, Fu S, Deng X. Targeted therapeutic strategies for triple-negative breast cancer. Front Oncol. 2021. https://doi.org/10.3389/fonc.2021.731535.
Zhu D, Shuichan Xu, Deyanat-Yazdi G, Peng SX, Barnes LA, Narla RK, Tran T, et al. Synthetic lethal strategy identifies a potent and selective TTK and CLK1/2 inhibitor for treatment of triple-negative breast cancer with a compromised G1–S checkpoint. Mol Cancer Ther. 2018;17(8):1727–38.
Jeong S. SR proteins: binders, regulators, and connectors of RNA. Mol Cells. 2017;40(1):1.
Yoshida T, Kim JH, Carver K, Ying Su, Weremowicz S, Mulvey L, Yamamoto S, et al. CLK2 is an oncogenic kinase and splicing regulator in breast cancer. Can Res. 2015;75(7):1516–26.
Xu Q, Yali Xu, Pan Bo, Liangcai Wu, Ren X, Zhou Y, Mao F, et al. TTK is a favorable prognostic biomarker for triple-negative breast cancer survival. Oncotarget. 2016;7(49):81815.
Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23(3):555–67.
Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol. 2018;15(9):564–76.
Erber J, Steiner JD, Isensee J, Lobbes LA, Toschka A, Beleggia F, Schmitt A, et al. Dual inhibition of GLUT1 and the ATR/CHK1 kinase axis displays synergistic cytotoxicity in KRAS-mutant cancer cellssynergistic interaction between GLUT1 and the ATR/CHK1 axis. Can Res. 2019;79(19):4855–68.
López-Vallejo F, Caulfield T, Martínez-Mayorga K, Giulianotti MA, Nefzi A, Houghten RA, Medina-Franco JL. Integrating virtual screening and combinatorial chemistry for accelerated drug discovery. Comb Chem High Throughput Screen. 2011;14(6):475–87.
Chari S, Dworkin I. The conditional nature of genetic interactions: the consequences of wild-type backgrounds on mutational interactions in a genome-wide modifier screen. PLoS Genet. 2013;9(8):e1003661.
Ryan CJ, Mehta I, Kebabci N, Adams DJ (2023) Targeting synthetic lethal paralogs in cancer. Trends Cancer. https://doi.org/10.1016/j.trecan.2023.02.002
Acknowledgements
The authors sincerely thank the management of Bethune College, Vellore Institute of Technology, Vellore, and the Council of Scientific and Industrial Research (CSIR), New Delhi, for providing all the necessary amenities required for the development of this computational work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest during the course of this work.
Research involving human participants and/or animals
This review article does not contain any animal studies or involve any human participation by any of the authors.
Informed consent
All participants provided informed consent prior to their participation.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chatterjee, P., Karn, R., Isaac, A.E. et al. Unveiling the vulnerabilities of synthetic lethality in triple-negative breast cancer. Clin Transl Oncol 25, 3057–3072 (2023). https://doi.org/10.1007/s12094-023-03191-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03191-9