Abstract
Background
This study aims to genomically characterize melanoma of unknown primary (MUP) in comparison to melanomas of cutaneous primary (MCP).
Methods
Eligible cases were collected from the MSK-IMPACT™ Clinical Sequencing Cohort published in the cBioPortal database. Genomic analysis was performed using a hybridization-capture-based next-generation sequencing assay designed to detect mutations, small insertions and deletions, copy number alterations, and genomic rearrangements.
Results
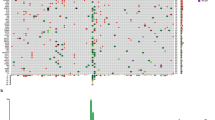
Among 462 patients of whom 18.4% had MUP, brain metastasis was more common among patients with MUP (23% vs 7.1%). The differences in genomic profiling between MCP and MUP did not reach statistical significance. The 187 MCP and 44 MUP patients treated with immune checkpoint inhibitors had a median overall survival of 49 and 44 months, respectively (p = 0.705).
Conclusions
The differences in somatic mutation patterns and survival outcomes were not statistically significant. These findings may allude to similar carcinogenic processes but should be considered exploratory and interpreted with caution.





Similar content being viewed by others
Data availability
The data are publically available on https://www.cbioportal.org/.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.
Kamposioras K, Pentheroudakis G, Pectasides D, Pavlidis N. Malignant melanoma of unknown primary site. To make the long story short. A systematic review of the literature. Crit Rev Oncol Hematol. 2011;78:112–26.
Lee CC, Faries MB, Wanek LA, Morton DL. Improved survival for stage IV melanoma from an unknown primary site. J Clin Oncol. 2009;27:3489–95.
Dasgupta T, Bowden L, Berg JW. Malignant melanoma of unknown primary origin. Surg Gynecol Obstet. 1963;117:341–5.
Rassy E, Pavlidis N. Progress in refining the clinical management of cancer of unknown primary in the molecular era. Nat Rev Clin Oncol. 2020;17:541–54.
Rassy E, Assi T, Pavlidis N. Exploring the biological hallmarks of cancer of unknown primary: where do we stand today? Br J Cancer. 2020;122:1124–32.
Verver D, van der Veldt A, van Akkooi A, Verhoef C, Grünhagen DJ, Louwman WJ. Treatment of melanoma of unknown primary in the era of immunotherapy and targeted therapy: a Dutch population-based study. Int J Cancer. 2020;146:26–34.
Gos A, Jurkowska M, van Akkooi A, Robert C, Kosela-Paterczyk H, Koljenović S, et al. Molecular characterization and patient outcome of melanoma nodal metastases and an unknown primary site. Ann Surg Oncol. 2014;21:4317–23.
Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13.
Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6.
Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn JMD. 2015;17:251–64.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Rabbie R, Ferguson P, Wong K, Couturier D-L, Moran U, Turner C, et al. The mutational landscape of melanoma brain metastases presenting as the first visceral site of recurrence. Br J Cancer. 2021;124:156–60.
Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9.
Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61.
Kiuru M, Busam KJ. The NF1 gene in tumor syndromes and melanoma. Lab Investig J Tech Methods Pathol. 2017;97:146–57.
Cirenajwis H, Lauss M, Ekedahl H, Törngren T, Kvist A, Saal LH, et al. NF1-mutated melanoma tumors harbor distinct clinical and biological characteristics. Mol Oncol. 2017;11:438–51.
Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002.
Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63.
Whittaker SR, Theurillat J-P, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–62.
Wilson MA, Zhao F, Khare S, Roszik J, Woodman SE, D’Andrea K, et al. Copy number changes are associated with response to treatment with carboplatin, paclitaxel, and sorafenib in melanoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2016;22:374–82.
Flores JF, Walker GJ, Glendening JM, Haluska FG, Castresana JS, Rubio MP, et al. Loss of the p16INK4a and p15INK4b genes, as well as neighboring 9p21 markers, in sporadic melanoma. Cancer Res. 1996;56:5023–32.
Haluska FG, Hodi FS. Molecular genetics of familial cutaneous melanoma. J Clin Oncol Off J Am Soc Clin Oncol. 1998;16:670–82.
Greshock J, Nathanson K, Medina A, Ward MR, Herlyn M, Weber BL, et al. Distinct patterns of DNA copy number alterations associate with BRAF mutations in melanomas and melanoma derived cell lines. Genes Chromosomes Cancer. 2009;48:419–28.
Potrony M, Badenas C, Aguilera P, Puig-Butille JA, Carrera C, Malvehy J, et al. Update in genetic susceptibility in melanoma. Ann Transl Med [Internet]. 2015;3. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4583600/ [cited 2021 Jan 12].
Bae JM, Choi YY, Kim DS, Lee JH, Jang HS, Lee JH, et al. Metastatic melanomas of unknown primary show better prognosis than those of known primary: a systematic review and meta-analysis of observational studies. J Am Acad Dermatol. 2015;72:59–70.
Gambichler T, Chatzipantazi M, Schröter U, Stockfleth E, Gedik C. Patients with melanoma of unknown primary show better outcome under immune checkpoint inhibitor therapy than patients with known primary: preliminary results. Oncoimmunology. 2019;8:e1677139.
Ellebaek E, Bastholt L, Schmidt H, Svane IM, Donia M. The real-world outcome of metastatic melanoma: unknown primary vs. known cutaneous. Int J Cancer. 2019;145:3173–4.
Boussios S, Rassy E, Samartzis E, Moschetta M, Sheriff M, Pérez-Fidalgo JA, et al. Melanoma of unknown primary: new perspectives for an old story. Crit Rev Oncol Hematol. 2020;158:103208.
Funding
None.
Author information
Authors and Affiliations
Contributions
Study concept and design: ER and NP; data acquisition: ER and AC; data analysis and interpretation: ER, AC and SB; statistical analysis: ER; manuscript editing and critical review: SB, CF, JK, and NP.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The data from the cBioPortal database do not require ethical approval.
Informed consent
The datasets are publically available. there was not any interactions with the patients who remain anyonymous on the cbioportal plateform.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rassy, E., Boussios, S., Chebly, A. et al. Comparative genomic characterization of melanoma of known and unknown primary. Clin Transl Oncol 23, 2302–2308 (2021). https://doi.org/10.1007/s12094-021-02629-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-021-02629-2