Abstract
Wolbachia is an obligate intracellular bacterium with a high frequency of infection and a continental distribution in arthropods and nematodes. This endosymbiont can induce various reproductive phenotypes in their hosts and has been previously found naturally in several pests including thrips (Thripidae). These insects cause physical fruit damage and economic losses in avocado. The presence of Wolbachia was evaluated for the first time in avocado thrips populations of Frankliniella sp. and Scirtothrips hansoni sp.n. from eastern Antioquia. DNA from adult thrips individuals was used to assess the detection of Wolbachia by amplifying a fragment (600 bp) of the Wolbachia major surface protein (wsp) gene. Results confirmed the presence of two new Wolbachia strains in these two thrips species, with a higher percentage of natural infection in S. hansoni sp.n. The first Wolbachia species was found in Frankliniella sp. and belongs to supergroup A and the second was detected in S. hansoni sp.n. and is part of supergroup B. Wolbachia was more frequently found in females (32.73%), and only found in one male. Analysis of phylogenetic relationships, suggests that the two new Wolbachia sequences (wFran: Frankliniella and wShan: Scirtothrips hansoni) detected here represent two new groups for this endosymbiont. The haplotype network shows the presence of two possible haplotypes for each strain. Future studies to evaluate the possible use of Wolbachia as a control agent in avocado thrips are necessary.

Similar content being viewed by others
Data Availability
The datasets supporting the conclusions of this article are included within the article and its additional file. The newly-generated sequences are submitted to the GenBank database under accession numbers MT649512-MT649530 for 16S rRNA sequence and MT647247-MT647265 for wsp sequence.
References
Li K, Stanojević M, Stamenković G et al (2019) Insight into diversity of bacteria belonging to the order Rickettsiales in 9 arthropods species collected in Serbia. Sci Rep 9:18680. https://doi.org/10.1038/s41598-019-55077-y
Landmann F (2019) The Wolbachia Endosymbionts. Bact Intracellularity. https://doi.org/10.1128/microbiolspec.bai-0018-2019
Tolley SJA, Nonacs P, Sapountzis P (2019) Wolbachia horizontal transmission events in ants: What do we know and what can we learn? Front Microbiol 10:296. https://doi.org/10.3389/fmicb.2019.00296
Vivero RJ, Cadavid-Restrepo G, Moreno Herrera CX, Uribe Soto SI (2017) Molecular detection and identification of Wolbachia in three species of the genus Lutzomyia on the Colombian Caribbean coast. Parasit Vectors 10:110. https://doi.org/10.1186/s13071-017-2031-x
Inácio da Silva LM, Dezordi FZ, Paiva MHS, Wallau GL (2021) Systematic review of Wolbachia symbiont detection in mosquitoes: an entangled topic about methodological power and true symbiosis. Pathogens 10:1–20. https://doi.org/10.3390/pathogens10010039
Baldo L, Werren JH (2007) Revisiting Wolbachia supergroup typing based on WSP: spurious lineages and discordance with MLST. Curr Microbiol 55:81–87. https://doi.org/10.1007/s00284-007-0055-8
Pascar J, Chandler CH (2018) A bioinformatics approach to identifying Wolbachia infections in arthropods. PeerJ 6:e5486. https://doi.org/10.7717/peerj.5486
Bravo-Pérez D, Santillán-Galicia MT, Johansen-Naime RM et al (2018) Species diversity of thrips (Thysanoptera) in selected avocado orchards from Mexico based on morphology and molecular data. J Integr Agric 17:2509–2517. https://doi.org/10.1016/S2095-3119(18)62044-1
Varon Devia EH (2014) Trips, bichos candela. In: Actualización tecnológica y buenas prácticas agrícolas (BPA) en el cultivo de aguacate. Manual Técnico CORPOICA, Centro de Investigación la Selva. Corporación Colombiana de Investigación Agropecuaria, Antioquia, pp 257–265. http://hdl.handle.net/20.500.12324/12616
Arakaki N, Miyoshi T, Noda H (2001) Wolbachia-mediated parthenogenesis in the predatory thrips Franklinothrips vespiformis (Thysanoptera: Insecta). Proc R Soc B Biol Sci 268:1011–1016. https://doi.org/10.1098/rspb.2001.1628
Ding T, Chi H, Gökçe A et al (2018) Demographic analysis of arrhenotokous parthenogenesis and bisexual reproduction of Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae). Sci Rep 8:3346. https://doi.org/10.1038/s41598-018-21689-z
Dickey AM, Trease AJ, Jara-Cavieres A et al (2014) Estimating bacterial diversity in Scirtothrips dorsalis (Thysanoptera: Thripidae) via next generation sequencing. Florida Entomol 97:362–366. https://doi.org/10.1896/054.097.0204
Powell CM, Lopez Montiel A, Beddingfield B et al (2015) Comparison of bacterial communities of flower thrips (Frankliniella tritici) and potato psyllid (Bactericera cockerelli). Southwest Entomol 40:765–773. https://doi.org/10.3958/059.040.0404
Kaczmarczyk A, Kucharczyk H, Kucharczyk M et al (2018) First insight into microbiome profile of fungivorous thrips Hoplothrips carpathicus (Insecta: Thysanoptera) at different developmental stages: molecular evidence of Wolbachia endosymbiosis. Sci Rep 8:14376. https://doi.org/10.1038/s41598-018-32747-x
Gawande SJ, Anandhan S, Ingle A et al (2019) Microbiome profiling of the onion thrips, Thrips tabaci Lindeman (Thysanoptera: Thripidae). PLoS ONE 14:e0223281. https://doi.org/10.1371/journal.pone.0223281
Cavalleri A, Mound LA (2012) Toward the identification of Frankliniella species in Brazil (Thysanoptera, Thripidae). Zootaxa 30:1–30. https://doi.org/10.5281/zenodo.246160
Mound L, Hoddle M (2016) The Scirtothrips perseae species-group (Thysanoptera), with one new species from avocado, Persea americana. Zootaxa 4079:388–392. https://doi.org/10.11646/zootaxa.4079.3.7
Cano-Calle D, Saldamando-Benjumea CI, Moreno-Herrera CX, Arango-Isaza RE (2020) Molecular characterization of thrips (Thysanoptera: Thripidae) from commercial avocado crops (Persea americana Mill) in eastern Antioquia and study of the associated microbial diversity. Universidad Nacional de Colombia. https://repositorio.unal.edu.co/handle/unal/78594
Cole JR, Wang Q, Cardenas E et al (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:141–145. https://doi.org/10.1093/nar/gkn879
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:113. https://doi.org/10.1186/1471-2105-5-113
Martin DP, Murrell B, Golden M et al (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. https://doi.org/10.1093/ve/vev003
Ronquist F, Teslenko M, Van Der Mark P et al (2012) Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC et al (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302
Bykov RA, Yudina MA, Gruntenko NE et al (2019) Prevalence and genetic diversity of Wolbachia endosymbiont and mtDNA in Palearctic populations of Drosophila melanogaster. BMC Evol Biol 19:48. https://doi.org/10.1002/arch.21776
Jacob TK, Dsilva S, Senthil Kumar CM et al (2014) Single strain infection of adult and larval cardamom thrips (Sciothrips cardamomi) by Wolbachia subgroup Con belonging to supergroup B in India. Invertebr Reprod Dev 59:1–8. https://doi.org/10.1080/07924259.2014.970237
Karami M, Moosa-Kazemi SH, Oshaghi MA et al (2016) Wolbachia endobacteria in natural populations of Culex pipiens of Iran and its phylogenetic congruence. J Arthropod Borne Dis 10:347–363
Saurav G, Daimei G, Singh VR et al (2016) Detection and Localization of Wolbachia in Thrips palmi Karny (Thysanoptera: Thripidae). Indian J Microbiol. https://doi.org/10.1007/s12088-016-0567-7
Ambika S, Rajagopal R (2018) Lumen anatomy and localization of Wolbachia sp. in the thrips, Plicothrips apicalis (Bagnall). Curr Sci 115:1297–1304. https://doi.org/10.18520/cs/v115/i7/1297-1304
Ford SA, Allen SL, Ohm JR et al (2019) Selection on Aedes aegypti alters Wolbachia-mediated dengue virus blocking and fitness. Nat Microbiol 4:1832–1839. https://doi.org/10.1038/s41564-019-0533-3
Liu H, Li H, Song F et al (2017) Novel insights into mitochondrial gene rearrangement in thrips (Insecta: Thysanoptera) from the grass thrips, Anaphothrips obscurus. Sci Rep 7:4284. https://doi.org/10.1038/s41598-017-04617-5
Braig HR, Zhou W, Dobson SL, Neill SLO (1998) Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J Bacteriol 180:2373–2378
Yang XH, Zhu DH, Liu Z et al (2013) High levels of multiple infections, recombination and horizontal transmission of Wolbachia in the Andricus mukaigawae (Hymenoptera; Cynipidae) communities. PLoS ONE 8:e78970. https://doi.org/10.1371/journal.pone.0078970
van der Kooi CJ, Schwander T (2014) Evolution of asexuality via different mechanisms in grass thrips (Thysanoptera: Aptinothrips). Evolution 68:1883–1893. https://doi.org/10.1111/evo.12402
Acknowledgements
We are thankful to Mr. Roberto Aguilar and the laboratorio de Biología Celular y Molecular of the Universidad Nacional de Colombia, sede Medellín. We thank the owners of the farms who permitted the collection of biological material.
Funding
The research reported in this publication was supported by the Universidad Nacional de Colombia under Project Code 202010013471, contract number FP44842-132-2017. This material is based on work supported by the Ministry of science, technology, and innovation (Minciencias) of Colombia Graduate Research Fellowship Program for Daniela Cano-Calle No.727 in 2015 under Grant No. 201010020475. Foundation for the Promotion of Research and Technology, Bank of the Republic of Colombia Project No 201423.
Author information
Authors and Affiliations
Contributions
All authors contributed to the writing and revision of the manuscript. Cano-Calle D. contributed to the development of experiments in the laboratory and field trips. Vivero-Gomez R., Moreno-Herrera C., Saldamando-Benjumea C., and Arango-Isaza R., Designed the study, analyzed the data, and contributed to writing the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that no conflict of interest exists in the study.
Ethics Statement
The insect collections were made between April and August 2017 through the certificate issued by Universidad Nacional de Colombia, Sede Medellín with the permission to collect specimens of wild species of biological diversity with scientific research purposes produced by the National Authority for Environmental Licenses (ANLA) to Universidad Nacional de Colombia through resolution No. 0255 of March 14, 2014 (article 3). Thrips were collected on private property and permission was received from the landowners prior to sampling procedures.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
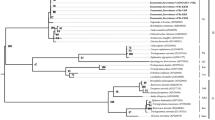
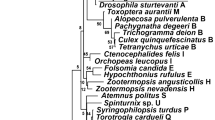
Below is the link to the electronic supplementary material. Fig. 1 Map of the localities studied in Colombia. Fig. 2 PCR amplification from Wolbachia of the16S rRNA and wsp genes from wild thrips. a) amplification of 16S rRNA gene and b) amplification of wsp gene. Lines 1-15 correspond to Scirtothrips hansoni sp.n.. C-: negative control (mix PCR plus sterile water free DNAse and RNAse); C +: positive control; M: 100 bp molecular marker. Fig. 3 Wolbachia phylogeny found in populations of natural thrips. a) Maximum likelihood Tree using partial sequences of 16S rRNA gene from Wolbachia and Kimura 2 parameter with 1000 replicates. The numbers in nodes correspond to the bootstrap support inferred by analysis. Blue boxes correspond to the Wolbachia wShan strain found in Scirtothrips hansoni sp.n.and red boxes correspond to Wolbachia wFran strain from Frankliniella. Outgroups: Anaplasma marginale and Ehrlichia ruminantium. Fig. 4 Bayesian probability inference tree based on partial sequences of wsp gene obtained from GenBank and Wolbachia sequences amplified from avocado thrips. The posterior probability values are indicated by numbers on nodes. In blue are Wolbachia found in Scirtothrips hansoni and in red are those found in Frankliniella. Outgroup: Bemicia tabaci wBtab. Fig. 5 Wolbachia strain haplotype network. wShan and wFran strains correspond to Wolbachia strains found in this work in Scirtothrips hansoni sp. n. and Frankliniella, respectively. Blue and red boxes enclose two supergroups A and B with their haplotypes





12088_2021_951_MOESM6_ESM.docx
Table 1 Information on samples used for the study. * Possibly corresponds to F. gardeniae or F. gossypiana. Supplementary file6 (DOCX 14 kb)
12088_2021_951_MOESM7_ESM.docx
Table 2 Frequency of natural infection by Wolbachia in thrips obtained from farms in eastern Antioquia, Colombia. Supplementary file7 (DOCX 12 kb)
Rights and permissions
About this article
Cite this article
Cano-Calle, D., Saldamando-Benjumea, C.I., Vivero-Gómez, R.J. et al. Two New Strains of Wolbachia Affecting Natural Avocado Thrips. Indian J Microbiol 61, 348–354 (2021). https://doi.org/10.1007/s12088-021-00951-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-021-00951-5