Abstract
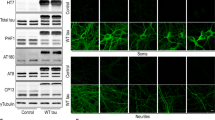
Proteasomal degradation of intrinsically disordered proteins, such as tau, is a critical component of proteostasis in both aging and neurodegenerative diseases. In this study, we investigated proteasomal activation by MK886 (MK). We previously identified MK as a lead compound capable of modulating tau oligomerization in a cellular FRET assay and rescuing P301L tau-induced cytotoxicity. We first confirmed robust proteasomal activation by MK using 20S proteasomal assays and a cellular proteasomal tau-GFP cleavage assay. We then show that MK treatment can significantly rescue tau-induced neurite pathology in differentiated SHSY5Y neurospheres. Due to this compelling result, we designed a series of seven MK analogs to determine if proteasomal activity is sensitive to structural permutations. Using the proteasome as the primary MOA, we examined tau aggregation, neurite outgrowth, inflammation, and autophagy assays to identify two essential substituents of MK that are required for compound activity: (1) removal of the N-chlorobenzyl group from MK negated both proteasomal and autophagic activity and reduced neurite outgrowth; and (2) removal of the indole-5-isopropyl group significantly improved neurite outgrowth and autophagy activity but reduced its anti-inflammatory capacity. Overall, our results suggest that the combination of proteasomal/autophagic stimulation and anti-inflammatory properties of MK and its derivatives can decrease tau-tau interactions and help rebalance dysfunctional proteostasis. Further development of MK to optimize its proteasomal, autophagic, and anti-inflammatory targets may lead to a novel therapeutic that would be beneficial in aging and neurodegenerative diseases.





Similar content being viewed by others
Data Availability
The datasets generated and/or analyzed during the current study are available upon request.
Abbreviations
- Aβ:
-
amyloid-beta
- AD:
-
Alzheimer’s disease
- BafA:
-
Bafilomycin A1
- BTZ:
-
Bortezomib
- EV:
-
empty vector
- 5-LO:
-
5-lipoxygenase
- FLAP:
-
5-lipoxygenase-activating protein
- FLT:
-
fluorescence lifetime
- FLT-FRET:
-
Fluorescence LifeTime-Förster Resonance Energy Transfer
- IDP:
-
intrinsically disordered protein
- MK:
-
MK-886
- MOA:
-
mechanism of action
- SHNS:
-
SHSY5Y neurosphere
- WT:
-
wildtype
References
Hipp MS, Kasturi P, Hartl FU (2019) The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol 20(7):421–435. https://doi.org/10.1038/s41580-019-0101-y
Lee MJ, Lee JH, Rubinsztein DC (2013) Tau degradation: the ubiquitin-proteasome system versus the autophagy-lysosome system. Prog Neurobiol 105:49–59. https://doi.org/10.1016/j.pneurobio.2013.03.001
Mazzulli JR, Zunke F, Isacson O, Studer L, Krainc D (2016) α-Synuclein-induced lysosomal dysfunction occurs through disruptions in protein trafficking in human midbrain synucleinopathy models. Proc Natl Acad Sci U S A 113(7):1931–1936. https://doi.org/10.1073/pnas.1520335113
Robak LA, Jansen IE, van Rooij J, Uitterlinden AG, Kraaij R, Jankovic J, Heutink P, Shulman JM et al (2017) Excessive burden of lysosomal storage disorder gene variants in Parkinson’s disease. Brain 140(12):3191–3203. https://doi.org/10.1093/brain/awx285
Sweeney P, Park H, Baumann M, Dunlop J, Frydman J, Kopito R, McCampbell A, Leblanc G et al (2017) Protein misfolding in neurodegenerative diseases: implications and strategies. Transl Neurodegener 6:6. https://doi.org/10.1186/s40035-017-0077-5
Chen JJ, Nathaniel DL, Raghavan P, Nelson M, Tian R, Tse E, Hong JY, See SK et al (2019) Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J Biol Chem 294(50):18952–18966. https://doi.org/10.1074/jbc.RA119.009432
Congdon EE, Sigurdsson EM (2018) Tau-targeting therapies for Alzheimer disease. Nat Rev Neurol 14(7):399–415. https://doi.org/10.1038/s41582-018-0013-z
Njomen E, Tepe JJ (2019) Proteasome activation as a new therapeutic approach to target proteotoxic disorders. J Med Chem 62(14):6469–6481. https://doi.org/10.1021/acs.jmedchem.9b00101
Cheng C, Reis SA, Adams ET, Fass DM, Angus SP, Stuhlmiller TJ, Richardson J, Olafson H et al (2021) High-content image-based analysis and proteomic profiling identifies tau phosphorylation inhibitors in a human iPSC-derived glutamatergic neuronal model of tauopathy. Sci Rep 11(1):17029. https://doi.org/10.1038/s41598-021-96227-5
Lo CH, Lim CK, Ding Z, Wickramasinghe SP, Braun AR, Ashe KH, Rhoades E, Thomas DD et al (2019) Targeting the ensemble of heterogeneous tau oligomers in cells: a novel small molecule screening platform for tauopathies. Alzheimers Dement 15(11):1489–1502. https://doi.org/10.1016/j.jalz.2019.06.4954
Rouzer CA, Ford-Hutchinson AW, Morton HE, Gillard JW (1990) MK886, a potent and specific leukotriene biosynthesis inhibitor blocks and reverses the membrane association of 5-lipoxygenase in ionophore-challenged leukocytes. J Biol Chem 265(3):1436–1442
Dixon RA, Diehl RE, Opas E, Rands E, Vickers PJ, Evans JF, Gillard JW, Miller DK (1990) Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature 343(6255):282–284. https://doi.org/10.1038/343282a0
Gillard J, Ford-Hutchinson AW, Chan C, Charleson S, Denis D, Foster A, Fortin R, Leger S et al (1989) L-663,536 (MK-886) (3-[1-(4-chlorobenzyl)-3-t-butyl-thio-5-isopropylindol-2-yl]-2,2 - dimethylpropanoic acid), a novel, orally active leukotriene biosynthesis inhibitor. Can J Physiol Pharmacol 67(5):456–464. https://doi.org/10.1139/y89-073
Miller DK, Gillard JW, Vickers PJ, Sadowski S, Léveillé C, Mancini JA, Charleson P, Dixon RA et al (1990) Identification and isolation of a membrane protein necessary for leukotriene production. Nature 343(6255):278–281. https://doi.org/10.1038/343278a0
Valera E, Dargusch R, Maher PA, Schubert D (2013) Modulation of 5-lipoxygenase in proteotoxicity and Alzheimer’s disease. J Neurosci 33(25):10512–10525. https://doi.org/10.1523/JNEUROSCI.5183-12.2013
Ben-Nissan G, Vimer S, Tarnavsky M, Sharon M (2019) Structural mass spectrometry approaches to study the 20S proteasome. Methods Enzymol 619:179–223. https://doi.org/10.1016/bs.mie.2018.12.029
Zhao X, Yang J (2010) Amyloid-β peptide is a substrate of the human 20S proteasome. ACS Chem Neurosci 1(10):655–660. https://doi.org/10.1021/cn100067e
Trader DJ, Simanski S, Dickson P (1861) Kodadek T (2017) Establishment of a suite of assays that support the discovery of proteasome stimulators. Biochim Biophys Acta Gen Subj 4:892–899. https://doi.org/10.1016/j.bbagen.2017.01.003
Coleman RA, Trader DJ (2019) Methods to discover and evaluate proteasome small molecule stimulators. Molecules 24(12). https://doi.org/10.3390/molecules24122341
Schmidt SJ, Garnes KT, Heys JR, Landvatter SW, Adams JL (1991) Synthesis of tritium labeled MK-886, a leukotriene biosynthesis inhibitor; employment of epibromohydrin as a masked electrophilic acetone synthon. J Label Compd Radiopharm 29(8):891–902. https://doi.org/10.1002/jlcr.2580290805
David DC, Layfield R, Serpell L, Narain Y, Goedert M, Spillantini MG (2002) Proteasomal degradation of tau protein. J Neurochem 83(1):176–185. https://doi.org/10.1046/j.1471-4159.2002.01137.x
Coleman RA, Mohallem R, Aryal UK, Trader DJ (2021) Protein degradation profile reveals dynamic nature of 20S proteasome small molecule stimulation. RSC Chem Biol 2(2):636–644. https://doi.org/10.1039/d0cb00191k
Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL (2007) Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell 27(5):731–744. https://doi.org/10.1016/j.molcel.2007.06.033
Liu CW, Corboy MJ, DeMartino GN, Thomas PJ (2003) Endoproteolytic activity of the proteasome. Science 299(5605):408–411. https://doi.org/10.1126/science.1079293
Nonaka T, Hasegawa M (2009) A cellular model to monitor proteasome dysfunction by alpha-synuclein. Biochemistry 48(33):8014–8022. https://doi.org/10.1021/bi900619j
La E, Kern JC, Atarod EB, Kehrer JP (2003) Fatty acid release and oxidation are factors in lipoxygenase inhibitor-induced apoptosis. Toxicol Lett 138(3):193–203. https://doi.org/10.1016/s0378-4274(02)00407-1
Ghosh J, Myers CE (1998) Inhibition of arachidonate 5-lipoxygenase triggers massive apoptosis in human prostate cancer cells. Proc Natl Acad Sci 95(22):13182–13187. https://doi.org/10.1073/pnas.95.22.13182
Dittmann KH, Mayer C, Rodemann HP, Petrides PE, Denzlinger C (1998) MK-886, a leukotriene biosynthesis inhibitor, induces antiproliferative effects and apoptosis in HL-60 cells. Leuk Res 22(1):49–53. https://doi.org/10.1016/s0145-2126(97)00132-x
Klegeris A, McGeer P (2003) Toxicity of human monocytic THP-1 cells and microglia toward SH-SY5Y neuroblastoma cells is reduced by inhibitors of 5-lipoxygenase and its activating protein FLAP Abstract. J Leukoc Biol 73(3):369–378. https://doi.org/10.1189/jlb.1002482
Seidel D, Krinke D, Jahnke HG, Hirche A, Kloß D, Mack TG, Striggow F, Robitzki A (2012) Induced tauopathy in a novel 3D-culture model mediates neurodegenerative processes: a real-time study on biochips. PLoS One 7(11):e49150. https://doi.org/10.1371/journal.pone.0049150
Harris G, Hogberg H, Hartung T, Smirnova L (2017) 3D differentiation of LUHMES cell line to study recovery and delayed neurotoxic effects. Curr Protoc Toxicol 73:11.23.11–11.23.28. https://doi.org/10.1002/cptx.29
Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, Gallego C, Comella JX (2000) Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated neurotrophic factor-dependent human neuron-like cells. J Neurochem 75(3):991–1003. https://doi.org/10.1046/j.1471-4159.2000.0750991.x
Smirnova L, Harris G, Delp J, Valadares M, Pamies D, Hogberg HT, Waldmann T, Leist M et al (2016) A LUHMES 3D dopaminergic neuronal model for neurotoxicity testing allowing long-term exposure and cellular resilience analysis. Arch Toxicol 90(11):2725–2743. https://doi.org/10.1007/s00204-015-1637-z
Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, Mandelkow E (1998) Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer’s disease. J Cell Biol 143(3):777–794. https://doi.org/10.1083/jcb.143.3.777
Rosa E, Mahendram S, Ke YD, Ittner LM, Ginsberg SD, Fahnestock M (2016) Tau downregulates BDNF expression in animal and cellular models of Alzheimer’s disease. Neurobiol Aging 48:135–142. https://doi.org/10.1016/j.neurobiolaging.2016.08.020
Huang EJ, Reichardt LF (2001) Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci 24:677–736. https://doi.org/10.1146/annurev.neuro.24.1.677
Hofer MM, Barde YA (1988) Brain-derived neurotrophic factor prevents neuronal death in vivo. Nature 331(6153):261–262. https://doi.org/10.1038/331261a0
Dasgupta S, Castro LM, Dulman R, Yang C, Schmidt M, Ferro ES, Fricker LD (2014) Proteasome inhibitors alter levels of intracellular peptides in HEK293T and SH-SY5Y cells. PLoS One 9(7):e103604. https://doi.org/10.1371/journal.pone.0103604
Giguere CJ, Schnellmann RG (2008) Limitations of SLLVY-AMC in calpain and proteasome measurements. Biochem Biophys Res Commun 371(3):578–581. https://doi.org/10.1016/j.bbrc.2008.04.133
Guthrie CR, Kraemer BC (2011) Proteasome inhibition drives HDAC6-dependent recruitment of tau to aggresomes. J Mol Neurosci 45(1):32–41. https://doi.org/10.1007/s12031-011-9502-x
Verplank JJS, Lokireddy S, Zhao J, Goldberg AL (2019) 26S Proteasomes are rapidly activated by diverse hormones and physiological states that raise cAMP and cause Rpn6 phosphorylation. Proc Natl Acad Sci 116(10):4228–4237. https://doi.org/10.1073/pnas.1809254116
Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, Kodama A, Ishihara T et al (2016) An autophagic flux probe that releases an internal control. Mol Cell 64(4):835–849. https://doi.org/10.1016/j.molcel.2016.09.037
Zhu K, Dunner K, McConkey DJ (2010) Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 29(3):451–462. https://doi.org/10.1038/onc.2009.343
Dainichi T, Kabashima K, Ivanov II, Goto Y (2021) Editorial: regulation of Immunity by Non-Immune Cells. Front Immunol 12:770847. https://doi.org/10.3389/fimmu.2021.770847
Poirier SJ, Boudreau LH, Flamand N, Surette ME (2020) LPS induces ALOX5 promoter activation and 5-lipoxygenase expression in human monocytic cells. Prostaglandins Leukot Essent Fatty Acids 154:102078. https://doi.org/10.1016/j.plefa.2020.102078
Lin HC, Lin TH, Wu MY, Chiu YC, Tang CH, Hour MJ, Liou HC, Tu HJ et al 5-Lipoxygenase inhibitors attenuate TNF-α-induced inflammation in human synovial fibroblasts. PLoS One 9(9):e107890. https://doi.org/10.1371/journal.pone.0107890
Ukmar-Godec T, Fang P, Ibáñez de Opakua A, Henneberg F, Godec A, Pan KT, Cima-Omori MS, Chari A et al (2020) Proteasomal degradation of the intrinsically disordered protein tau at single-residue resolution. Sci Adv 6(30):eaba3916. https://doi.org/10.1126/sciadv.aba3916
Caballero B, Wang Y, Diaz A, Tasset I, Juste YR, Stiller B, Mandelkow EM, Mandelkow E et al (2018) Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell 17(1). https://doi.org/10.1111/acel.12692
Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC (2009) Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell 33(4):517–527. https://doi.org/10.1016/j.molcel.2009.01.021
Korolchuk VI, Menzies FM, Rubinsztein DC (2010) Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584(7):1393–1398. https://doi.org/10.1016/j.febslet.2009.12.047
Njomen E, Tepe JJ (2019) Regulation of autophagic flux by the 20S proteasome. Cell Chem Biol 26(9):1283–1294.e1285. https://doi.org/10.1016/j.chembiol.2019.07.002
Zhao J, Goldberg AL (2016) Coordinate regulation of autophagy and the ubiquitin proteasome system by MTOR. Autophagy 12(10):1967–1970. https://doi.org/10.1080/15548627.2016.1205770
Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, Jacobs AH, Wyss-Coray T et al (2015) Neuroinflammation in Alzheimer’s disease. Lancet Neurol 14(4):388–405. https://doi.org/10.1016/S1474-4422(15)70016-5
Giannopoulos PF, Joshi YB, Chu J, Praticò D (2013) The 12-15-lipoxygenase is a modulator of Alzheimer’s-related tau pathology in vivo. Aging Cell 12(6):1082–1090. https://doi.org/10.1111/acel.12136
Chu J, Praticò D (2011) 5-lipoxygenase as an endogenous modulator of amyloid β formation in vivo. Ann Neurol 69(1):34–46. https://doi.org/10.1002/ana.22234
Chu J, Praticò D (2011) Pharmacologic blockade of 5-lipoxygenase improves the amyloidotic phenotype of an Alzheimer’s disease transgenic mouse model involvement of γ-secretase. Am J Pathol 178(4):1762–1769. https://doi.org/10.1016/j.ajpath.2010.12.032
Chu J, Praticò D (2009) The 5-lipoxygenase as a common pathway for pathological brain and vascular aging. Cardiovasc Psychiatry Neurol 2009:174657. https://doi.org/10.1155/2009/174657
Chu J, Lauretti E, Di Meco A, Praticò D (2013) FLAP pharmacological blockade modulates metabolism of endogenous tau in vivo. Transl Psychiatry 3:e333. https://doi.org/10.1038/tp.2013.106
Vagnozzi AN, Giannopoulos PF, Praticò D (2018) Brain 5-lipoxygenase over-expression worsens memory, synaptic integrity, and tau pathology in the P301S mice. Aging Cell 17(1). https://doi.org/10.1111/acel.12695
Giannopoulos PF, Chu J, Joshi YB, Sperow M, Li JG, Kirby LG, Praticò D (2013) 5-lipoxygenase activating protein reduction ameliorates cognitive deficit, synaptic dysfunction, and neuropathology in a mouse model of Alzheimer’s disease. Biol Psychiatry 74(5):348–356. https://doi.org/10.1016/j.biopsych.2013.04.009
Vagnozzi AN, Giannopoulos PF, Praticò D (2017) The direct role of 5-lipoxygenase on tau pathology, synaptic integrity and cognition in a mouse model of tauopathy. Transl Psychiatry 7(12):1288. https://doi.org/10.1038/s41398-017-0017-2
Acknowledgements
We thank Noboru Mizushima for his generous gift (pMRX-IP-GFP-LC3-RFP-LC3dG) construct. (Addgene plasmid #84572; http://n2t.net/addgene:84572; RRID:Addgene 84572).
Funding
The authors disclose receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the U.S. National Institutes of Health (NIH) to J.N.S. (NINDS NS117968, NIA AG073734, and NIGMS GM131814).
Author information
Authors and Affiliations
Contributions
E.E.L directed the study, designed, and performed cellular experiments, and wrote the manuscript; MY synthesized the compounds and edited the manuscript; NNK performed the DLS experiments; NV assisted with DLS and experiments probing the inflammatory signaling pathway; ARB conducted FLPR experiments and edited the manuscript; DMF edited the manuscript; and J.N.S. conceived of the study and edited the manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
This research did not contain any studies involving human or animal participants.
Consent to Participate
This research did not contain any studies involving human or animal participants.
Consent for Publication
This research did not contain any studies involving human participants.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
Supplemental data includes detailed materials and methods as well as 1 Scheme, 11 Supplemental Figures, and 1 Supplemental Table. (DOCX 18.5 mb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liao, E.E., Yang, M., Nathan Kochen, N. et al. Proteasomal Stimulation by MK886 and Its Derivatives Can Rescue Tau-Induced Neurite Pathology. Mol Neurobiol 60, 6133–6144 (2023). https://doi.org/10.1007/s12035-023-03417-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03417-5