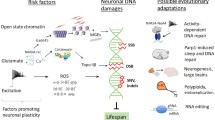
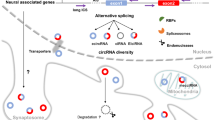
Abstract
We have previously outlined the main properties of brain metabolic DNA (BMD) and its involvement in circadian oscillations, learning, and post-trial sleep. The presence of BMD in certain subcellular fractions and their behavior in cesium gradients have suggested that BMD originates from cytoplasmic reverse transcription and subsequently acquires a double-stranded configuration. More recently, it has been reported that some DNA sequences of cytoplasmic BMD in learning mice are different from that of the control animals. Furthermore, BMD is located in vicinity of the genes involved in different modifications of synaptic activity, suggesting that BMD may contribute to the brain’s response to the changing environment. The present review outlines recent data with a special emphasis on reverse transcription of BMD that may recapitulate the molecular events at the time of the “RNA world” by activating mitochondrial telomerase and generating RNA templates from mitochondrial transcripts. The latter unexpected role of mitochondria is likely to promote a better understanding of mitochondrial contribution to cellular interactions and eukaryotic evolution. An initial step regards the role of human mitochondria in embryonic BMD synthesis, which is exclusively of maternal origin. In addition, mitochondrial transcripts involved in reverse transcription of BMD might possibly reveal unexpected features elucidating mitochondrial involvement in cancer events and neurodegenerative disorders.
Similar content being viewed by others
Data Availability
Not applicable.
References
Pelc SR (1968) Biological implications of DNA-turnover in higher organisms. Acta Histochem Suppl 8:441–452
Giuditta A, Rutigliano B (2018) Brain metabolic DNA in rat cytoplasm. Mol Neurobiol 55:7476–7486. https://doi.org/10.1007/s12035-018-0932-0
Prisco M, Casalino J, Cefaliello C, Giuditta A (2019) Brain metabolic DNA is reverse transcribed in cytoplasm: evidence by immunofluorescence analysis. Mol Neurobiol 56:6770–6776. https://doi.org/10.1007/s12035-019-1569-3
Giuditta A, Casalino J (2020) Sequences of reverse transcribed brain DNA are modified by learning. Front Mol Neurosci 13:57. https://doi.org/10.3389/fnmol.2020.00057
Koenig H (1958) An autoradiographic study of nucleic acid and protein turnover in the mammalian neuraxis. J Biophys Biochem Cytol 4:785–792. https://doi.org/10.1083/jcb.4.6.785
Giuditta A (1983) Role of DNA in brain activity. In: Lajtha A (ed) Handbook of Neurochemistry, vol 5. Plenum Press, New York, pp 251–276
Pelc SR (1962) Incorporation of tritiated thymidine in various organs of the mouse. Nature 193:793–795. https://doi.org/10.1038/193793a0
Pelc SR (1964) Labelling of DNA and cell division in so called non-dividing tissues. J Cell Biol 22:21–28. https://doi.org/10.1083/jcb.22.1.21
Pelc SR (1972) Metabolic DNA in ciliated protozoa, salivary gland chromosomes, and mammalian cells. Int Rev Cytol 32:327–355. https://doi.org/10.1016/s0074-7696(08)60344-7
Stroun M, Charles P, Anker P, Pelc SR (1967) Metabolic DNA in heart and skeletal muscle and in the intestine of mice. Nature 216:716–717. https://doi.org/10.1038/216716a0
Appleton TC, Pelc SR, Tarbit MH (1969) Formation and loss of DNA in intestinal epithelium. J Cell Sci 5:45–55
Harris G, Pelc SR (1970) Incorporation of [3H] thymidine into the spleens of intact mice during the immune response to sheep erythrocytes (SRC). Immunol 19:865–878
Pelc SR, Viola-Magni MP (1969) Decrease of labeled DNA in cells of the adrenal medulla after intermittent exposure to cold. J Cell Biol 42:460–468. https://doi.org/10.1083/jcb.42.2.460
Giuditta A, Grassi-Zucconi G, Sadile AG (2017) Brain metabolic DNA in memory processing and genome turnover. Rev Neurosci 28:21–30. https://doi.org/10.1515/revneuro-2016-0027
Reinis S (1970) Delayed learning deficit produced by hydroxylamine. Physiol Behav 5:253–256. https://doi.org/10.1016/0031-9384(70)90075-2
Reinis S (1971) Further study of the learning deficit produced by hydroxylamine. Physiol Behav 6:31–33. https://doi.org/10.1016/0031-9384(71)90009-6
Reinis S (1972) Autoradiographic study of 3-H-thymidine incorporation into brain DNA during learning. Physiol Chem Phys 4:391–397
Reinis S (1972) Hydroxylamine as an amnestic agent. Agents Actions 2:216–222. https://doi.org/10.1007/BF02087045
Reinis S, Lamble RW (1972) Labeling of brain DNA by 3-H-thymidine during learning. Physiol Chem Phys 4:335–338
Reinis S, Abbott J, Clarke JJ (1972) Brain DNA changes during learning studied by administration of 5-iodo-2’-deoxyuridine. Physiol Chem Phys 4:440–448
Reinis S (1975) Effects of hydroxylamine on the consequences of long-lasting administration of morphine in mice. I. Effect on the morphine tolerance. Arch Int Pharmacol 215(222):229
Reinis S (1975) Effects of hydroxylamine on the consequences of long-lasting administration of morphine in mice. II. Time course of the hydroxylamine effect on morphine tolerance. Arch Int Pharmacol 215:230–237
Reinis S (1975) Effect of hydroxylamine on the consequences of long-lasting administration of morphine in mice. III. Effect on preferred drinking of morphine solution. Arch Int Pharmacol 215:238–245
Reinis S (1975) Incorporation of (3H)thymidine into brain DNA after cerebellar damage. Pediatr Res 9:807–811. https://doi.org/10.1203/00006450-197511000-00001
Kinberlin RH, Anger HS (1969) DNA synthesis in the glial cells of scrapie-affected mouse brain. J Neurochem 16:543–548. https://doi.org/10.1111/j.1471-4159.1969.tb06853.x
Kimberin RH (1972) The nature of the increased rate of DNA synthesis in scrapie-affected mouse brain. J Neurochem 19:2767–2778. https://doi.org/10.1111/j.1471-4159.1972.tb03814.x
Kimberlin RH, Shirt DB, Collis SC (1974) The turnover of isotopically labelled DNA in vivo in developing, adult and scrapie-affected mouse brain. J Neurochem 23:241–248. https://doi.org/10.1111/j.1471-4159.1974.tb06940.x
Merits I, Cain J (1969) Rapid loss of labeled DNA from rat brain due to radiation damage. Biochim Biophys Acta 174:315–321. https://doi.org/10.1016/0005-2787(69)90256-1
Merits I, Cain J (1970) Loss of labelled DNA from rat brain following injections of precursors with high specific radioactivity. II. DNA labelled with 5-[131I]iodo-2’-deoxyuridine and 5-[82Br]bromo-2’-deoxyuridine. Biochim Biophys Acta 209:327–338. https://doi.org/10.1016/0005-2787(70)90731-8
Giuditta A, Libonati M, Packard A, Prozzo N (1971) Nuclear counts in the brain lobes of Octopus vulgaris as a function of body size. Brain Res 25:55–62. https://doi.org/10.1016/0006-8993(71)90566-x
De Marianis B, Giuditta A (1978) Separation of nuclei with different DNA content from the subesophageal lobe of octopus brain. Brain Res 154:134–136. https://doi.org/10.1016/0006-8993(78)91059-4
De Marianis B, Olmo E, Giuditta A (1979) Excess DNA in the nuclei of the subesophageal region of octopus brain. J Comp Neurol 186:293–300. https://doi.org/10.1002/cne.901860211
Giuditta A, De Marianis B, Sorrentino P (2017) Hyperdiploid DNA from octopus brain is enriched in AT sequences. Rendiconti dellìAccademia di Scienze Fisiche e Matematiche LXXXIV 5–16
Giuditta A, Abrescia P, Rutigliano B (1978) Effect of electroshock on thymidine incorporation into rat brain DNA. J Neurochem 31:983–987. https://doi.org/10.1111/j.1471-4159.1978.tb00137.x
Perrone-Capano C, D’Onofrio G, Giuditta A (1982) DNA turnover in rat cerebral cortex. J Neurochem 38:52–56. https://doi.org/10.1111/j.1471-4159.1982.tb10852.x
Grassi ZG, Carandente F, Menichini E, Belia S, Giuditta A (1988) Circadian rhythms of DNA content in brain and kidney: effects of environmental stimulation. Chronobiol 15:195–204
Grassi Zucconi G, Menichini E, Castigli E, Belia S, Giuditta A (1988) Circadian oscillations of DNA synthesis in rat brain. Brain Res 447:253–261. https://doi.org/10.1016/0006-8993(88)91127-4
Grassi Zucconi G, Crognale MC, Bassetti MA, Giuditta A (1990) Environmental stimuli modulate the circadian rhythm of [3H- methyl] thymidine incorporation into brain DNA of male rats. Behav Brain Res 41:103–110. https://doi.org/10.1016/0166-4328(90)90146-6
Giuditta A, Perrone Capano C, D’Onofrio G, Toniatti C, Menna T, Hydèn H (1986) Synthesis of rat brain DNA during acquisition of an appetitive task. Pharmacol Biochem Behav 25:651–658. https://doi.org/10.1016/0091-3057(86)90155-3
Scaroni R, Ambrosini MV, Principato GB, Federici F, Ambrosi G, Giuditta A (1983) Synthesis of brain DNA during acquisition of an active avoidance task. Physiol Behav 30:577–582. https://doi.org/10.1016/0031-9384(83)90224-x
Papa M, Pellicano MP, Cerbone A et al (1995) Immediate early genes and brain DNA remodeling in the Naples high and low-excitability rat lines following exposure to a spatial novelty. Brain Res Bull 37:111–118. https://doi.org/10.1016/0361-9230(94)00254-x
Ashapkin VV, Romanov GA, Tushmalova NA, Vanyushin BF (1983) Selective DNA synthesis in the rat brain induced by learning. Biokhimija 48:355–362
Ivashkina OI, Zots MA, Bezriadnov DV, Anokhin KV (2012) Increased 5’-bromo-2’-deoxyuridine incorporation in various brain structures following passive avoidance training in mice. Bull Ex Bio Me 154:171–173. https://doi.org/10.1007/s10517-012-1901-7
Komissarova NV, Tiunova AA, Anokhin KV (2010) Selective impairments to memory consolidation in chicks produced by 5’-iodo-2’-deoxyuridine. Neurosci Behav Physiol 40:215–223. https://doi.org/10.1007/s11055-009-9237-0
Efimova OI, Anokhin KV (2012) 5-Bromo-2’-deoxyuridine impairs long-term food aversion memory in edible snail. Bull Exp Biol Med 153:767–770. https://doi.org/10.1007/s10517-012-1822-5
Shevelkin AV, Efimova OI, Nikitin VP, Anokhin KV, Sherstnev VV (2012) Specific changes in c-fos expression and colocalization with DNA in identified neuronal nuclei of edible snail following neurotransmitter application. Bull Exp Biol Med 153:734–737. https://doi.org/10.1007/s10517-012-1813-6
Ambrosini MV, Mariucci G, Bruschelli G, Colarieti L, Giuditta A (1995) Sequential hypothesis of sleep function. V. Lengthening of post-trial SS episodes in reminiscent rats. Physiol Behav 58:1043–1049. https://doi.org/10.1016/0031-9384(95)00143-7
Mariucci G, Bruschelli G, Colarieti L, Gambelunghe C, Ambrosini MV (1998) Ital J Zool 65:311–314
Giuditta A, Ambrosini MV, Scaroni R, Chiurulla C, Sadile A (1985) Effect of sleep on cerebral DNA synthesized during shuttle-box avoidance training. Physiol Behav 34:769–778. https://doi.org/10.1016/0031-9384(85)90376-2
Ambrosini MV, Sadile AG, Gironi Carnevale UA, Mattiaccio M, Giuditta A (1988) The sequential hypothesis on sleep function. I. Evidence that the structure of sleep depends on the nature of the previous waking experience. Physiol Behav 43:325–337. https://doi.org/10.1016/0031-9384(88)90196-5
Ambrosini MV, Sadile AG, Gironi Carnevale UA, Mattiaccio M, Giuditta A (1988) The sequential hypothesis on sleep function. II. A correlative study between sleep variables and newly synthesized brain DNA. Physiol Behav 43:339–350. https://doi.org/10.1016/0031-9384(88)90197-7
Langella M, Colarieti L, Ambrosini MV, Giuditta A (1992) The sequential hypothesis of sleep function. IV. A correlative analysis of sleep variables in learning and non-learning rats. Physiol Behav 51:227–238. https://doi.org/10.1016/0031-9384(92)90135-o
Rutigliano B, Giuditta A (2015) The unexpected recovery of misplaced data on brain metabolic DNA. Rendiconti dellìAccademia di Scienze Fisiche e Matematiche LXXXII 99–106
Cefaliello C, Prisco M, Crispino M, Giuditta A (2019) DNA in squid synaptosomes. Mol Neurobiol 56:56–60. https://doi.org/10.1007/s12035-018-1071-3
Bregnard A, Knüsel A, Kuenzle C (1975) Are all the neuronal nuclei polyploid? Histochem 43:59–61. https://doi.org/10.1007/BF00490154
Bregnard A, Kuenzle CC, Ruch F (1977) Cytophotometric and autoradiographic evidence for post-natal DNA synthesis in neurons of the rat cerebral cortex. Exp Cell Res 107:151–157. https://doi.org/10.1016/0014-4827(77)90396-2
Kuenzle CC, Bregnard A, Hübscher U, Ruch F (1978) Extra DNA in forebrain cortical neurons. Exp Cell Res 113:151–160. https://doi.org/10.1016/0014-4827(78)90095-2
Hobi R, Studer M, Ruch F, Kuenzle CC (1984) The DNA content of cerebral cortex neurons. Determinations by cytophotometry and high performance liquid chromatography. Brain Res 305:209–219. https://doi.org/10.1016/0006-8993(84)90427-x
Bibbiani C, Viola-Magni MP (1975) Metabolic DNA in the hepatocyte nuclei in newborn rats. Histochem 43:63–72. https://doi.org/10.1007/BF00490155
Mirmiran M, Maas YGH, Ariagno RL (2003) Development of fetal and neonatal sleep and circadian rhythms. Sleep Med Rev 7:321–334. https://doi.org/10.1053/smrv.2002.0243
Grassi Zucconi G, Belia S, Franciolini F, Menichini E, Giuditta A (1984) Effect of paradoxical sleep deprivation on DNA synthesis in fetal rat brain. Intern J Develop Neurosci 2:585–590. https://doi.org/10.1016/0736-5748(84)90036-4
Grassi Zucconi G, Belia S, Menichini E, Castigli E, Giuditta A (1986) Paradoxical sleep deprivation of the mother enhances DNA synthesis in fetal rat brain: autoradiographic and biochemical evidence. Int J Dev Neurosci 4:169–178. https://doi.org/10.1016/0736-5748(86)90042-0
Marco A, Meharena HS, Dileep V et al (2020) Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat Neurosci 23:1606–1617. https://doi.org/10.1038/s41593-020-00717-0
You X, Vlatkovic I, Babic A et al (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18:603–610. https://doi.org/10.1038/nn.3975
van Rossum D, Verheijen BM, Pasterkamp RJ (2016) Circular RNAs: novel regulators of neuronal development. Front Mol Neurosci 9:74. https://doi.org/10.3389/fnmol.2016.00074
Yang Q, Wu J, Zhao J et al (2018) Circular RNA expression profiles during the differentiation of mouse neural stem cells. BMC Syst Biol 12(Suppl 8):128. https://doi.org/10.1186/s12918-018-0651-1
Suberbielle E, Sanchez PE, Kravitz AV et al (2013) Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-F. Nat Neurosci 16:613–621. https://doi.org/10.1038/nn.3356
Madabhushi R, Gao F, Pfenning AR et al (2015) Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161:1592–1605. https://doi.org/10.1016/j.cell.2015.05.032
Giuditta A, Grassi Zucconi G, Sadile A (2020). Brain metabolic DNA: recent evidence for a mitochondrial connection. Rev Neurosci 32(1):93–100. https://doi.org/10.1515/revneuro-2020-0050
Fedorov A, Fedorova L (2004) Introns: mighty elements from the RNA world. Mol Evol 59:718–721. https://doi.org/10.1007/s00239-004-2660-5
Talini G, Branciamore S, Gallori E (2011) Ribozymes: flexible molecular devices at work. Biochimie 93:1998–2005. https://doi.org/10.1016/j.biochi.2011.06.026
Neveu M, Kim H-J, Benner SA (2013) The “strong” RNA world hypothesis: fifty years old. Astrobiol 13:391–403. https://doi.org/10.1089/ast.2012.0868
Sankaran N (2016) The RNA world at thirty: a look back with its author. J Mol Evol 83:169–175. https://doi.org/10.1007/s00239-016-9767-3
Zhu XH, Qiao H, Du F et al (2012) Quantitative imaging of energy expenditure in human brain. Neuroimage 60:2107–2117. https://doi.org/10.1016/j.neuroimage.2012.02.013
Cardanho-Ramos C, Faria-Pereira A, Morais VA (2020) Orchestrating mitochondria in neurons: cytoskeleton as the conductor. Cytoskeleton (Hoboken) 77:65–75. https://doi.org/10.1002/cm.21585
Berglund AK, Navarrete C, Engqvist MK et al (2017) Nucleotide pools dictate the identity and frequency of ribonucleotide incorporation in mitochondrial DNA. PLoS Genet 16:13. https://doi.org/10.1371/journal.pgen.1006628
Moss CF, Dalla Rosa I, Hunt LE et al (2017) Aberrant ribonucleotide incorporation and multiple deletions in mitochondrial DNA of the murine MPV17 disease model. Nucleic Acids Res 45:12808–12815. https://doi.org/10.1093/nar/gkx1009
Cluett TJ, Akman G, Reyes A et al (2018) Transcript availability dictates the balance between strand-asynchronous and strand-coupled mitochondrial DNA replication. Nucleic Acids Res 46:10771–10781. https://doi.org/10.1093/nar/gky852
Cheng Y, Liu P, Zheng Q et al (2018) Mitochondrial trafficking and processing of telomerase RNA TERC. Cell Rep 4:2589–2595. https://doi.org/10.1016/j.celrep.2018.08.003
Zheng Q, Huang J, Wang G (2019) Mitochondria, telomeres and telomerase subunits. Front Cell Dev Biol 7:274. https://doi.org/10.3389/fcell.2019.00274
Stott RT, Kritsky O, L-H, (2021) Profiling DNA break sites and transcriptional changes in response to contextual fear learning. PLoS ONE 16(7):e0249691. https://doi.org/10.1371/journal.pone.0249691
Panfoli I, Ravera S, Bruschi M, Candiano G, Morelli A (2011) Proteomics unravels the exportability of mitochondrial respiratory chains. Expert Rev Proteomics 8:231–239. https://doi.org/10.1586/epr.11.1
Thakurela S, Garding A, Jung RB, Müller C, Goebbels S et al (2016) The transcriptome of mouse central nervous system myelin. Sci Rep 6:25828. https://doi.org/10.1038/srep25828
Ghosh T, Almeida RG, Zhao C, Gonzalez MG, Stott K et al (2022) A retroviral origin of vertebrate myelin bioRxiv. https://doi.org/10.1101/2022.01.24.477350
Cruz ACP, Ferrasa A, Muotri AR, Herai RH (2018) Frequency and association of mitochondrial genetic variants with neurological disorders. Mitochondrion 46:345–360. https://doi.org/10.1016/j.mito.2018.09.005
Cuperfain AB, Zhang ZL, Kennedy JL, Gonçalves VF (2018) The complex interaction of mitochondrial genetics and mitochondrial pathways in psychiatric disease. Mol Neuropsychiatry 4:52–69. https://doi.org/10.1159/000488031
Wei W, Pagnamenta AT, Gleadall N et al (2020) Nuclear-mitochondrial DNA segments resemble paternally inherited mitochondrial DNA in humans. Nature Commun 11:1740. https://doi.org/10.1038/s41467-020-15336-3
Mfossa ACM, Puthenparampil HT, Inalegwu A et al (2019) Exposure to ionizing radiation triggers prolonged changes in circular RNA abundance in the embryonic mouse brain and primary neurons. Cells 8:778. https://doi.org/10.3390/cells8080778
Li M-L, Wang W, Zi-B J (2021) Circular RNAs in the central nervous system. Front Mol Biosci 8:629593. https://doi.org/10.3389/fmolb.2021.629593
Acknowledgements
We thank Prof. Marianna Crispino, University of Naples Federico II, and Dr. Chun Jong Tai, Stazione Zoologica Anton Dohrn, Naples, for their helpful collaboration and comments to our manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors equally contributed to this study, read, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
All authors agreed.
Consent for Publication
All authors agreed.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Giuditta, A., Zucconi, G.G. & Sadile, A. Brain Metabolic DNA: A Long Story and Some Conclusions. Mol Neurobiol 60, 228–234 (2023). https://doi.org/10.1007/s12035-022-03030-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-03030-y