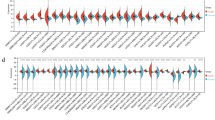
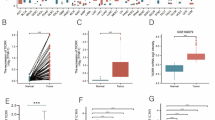
Abstract
Histological interpretation of the rare pleomorphic xanthoastrocytoma (PXA) has been the holy grail for treatment options. However, no stand-alone clinical interventions have been developed owing to the lack of gene expression profiling data in PXA/APXA patients. We first time report the comprehensive analyses of the coding as well as long non-coding RNA (lncRNA) signatures of PXA/APXA patients. Several genes such as IGFBP2, NF1, FOS, ERBB2, and lncRNAs such as NEAT1, HOTAIRM1, and GAS5 known to play crucial roles in glioma patients were also deregulated in PXA patients suggesting the commonality in the molecular signatures. PPI network, co-expression, and lncRNA-mRNA interaction studies unraveled hub genes (such as ERBB2, FOS, RPA1) and networks that may play a critical role in PXA biology. The most enriched pathways based on gene profiles were related to TLR, chemokine, MAPK, Rb, and PI3K-Akt signaling pathways. The lncRNA targets were enriched in glucuronidation, adipogenesis, TGF-beta signaling, EGF/EGFR signaling, and cell cycle pathways. Interestingly, several mRNAs like PARVG, and ABI2 were found to be targeted by multiple lncRNAs suggesting a tight control of their levels. Some of the most prominent lncRNA-mRNA pairs were LOC728730: MRPL9, XLOC_l2_011987: ASIC2, lnc-C1QTNF5-1: RNF26. Notably, several lncRNAs such as lnc-CETP-1, lnc-XRCC3-1, lnc-RPL31-1, lnc-USP13-1, and MAPKAPK5-AS1, and genes such as RPA1, NTRK3, and CNRP1 showed strong correlation to the progression-free survival of PXA patients suggesting their potential as novel biomarkers. Overall, the findings of this study may facilitate the development of a new realm of RNA biology in PXA that may have clinical significance in the future.








Similar content being viewed by others
Data Availability
The datasets analyzed during the current study are available in the GEO repository [GSE168904].
Code Availability
Not applicable.
References
Weller M, Wick W, Aldape K et al (2015) Glioma. Nat Rev Dis Prim 1(1):15017. https://doi.org/10.1038/nrdp.2015.17
Ostrom QT, Cioffi G, Gittleman H et al (2019) CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol 21(Supplement_5):v1–v100. https://doi.org/10.1093/neuonc/noz150
Watanabe N, Ishikawa E, Kohzuki H et al (2020) Malignant transformation of pleomorphic xanthoastrocytoma and differential diagnosis: case report. BMC Neurol 20(1):21. https://doi.org/10.1186/s12883-020-1601-2
Im S-H, Kee Chung C, Kim S-K, Cho B-K, Kim M-K, Chi JG (2004) Pleomorphic xanthoastrocytoma: a developmental glioneuronal tumor with prominent glioproliferative changes. J Neurooncol 66(1):17–27. https://doi.org/10.1023/B:NEON.0000013473.29458.9a
Macaulay RJB, Jay V, Hoffman HJ, Becker LE (1993) Increased mitotic activity as a negative prognostic indicator in pleomorphic xanthoastrocytoma: Case report. J Neurosurg 79(5):761–768. https://doi.org/10.3171/jns.1993.79.5.0761
Fouladi M, Jenkins J, Burger P et al (2001) Pleomorphic xanthoastrocytoma: favorable outcome after complete surgical resection. Neuro Oncol 3(3):184–192. https://doi.org/10.1093/neuonc/3.3.184
Crespo-Rodríguez AM, Smirniotopoulos JG, Rushing EJ (2007) MR and CT imaging of 24 pleomorphic xanthoastrocytomas (PXA) and a review of the literature. Neuroradiology 49(4):307–315. https://doi.org/10.1007/s00234-006-0191-z
Ida CM, Rodriguez FJ, Burger PC et al (2015) Pleomorphic xanthoastrocytoma: natural history and long-term follow-up. Brain Pathol 25(5):575–586. https://doi.org/10.1111/bpa.12217
Schindler G, Capper D, Meyer J et al (2011) Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol 121(3):397–405. https://doi.org/10.1007/s00401-011-0802-6
Freije WA, Castro-Vargas FE, Fang Z et al (2004) Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 64(18):6503 LP – 6510. https://doi.org/10.1158/0008-5472.CAN-04-0452
Ling H, Vincent K, Pichler M et al (2015) Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 34(39):5003–5011. https://doi.org/10.1038/onc.2014.456
Chen W, Xu X-K, Li J-L, et al. (2017) MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget;8(14).. https://www.oncotarget.com/article/15199/text/.
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15(suppl_1):R17–R29. https://doi.org/10.1093/hmg/ddl046
Amaral PP, Dinger ME, Mercer TR, Mattick JS (2008) The eukaryotic genome as an RNA machine. Science (80) 319(5871):1787 LP – 1789. https://doi.org/10.1126/science.1155472
Carninci P, Kasukawa T, Katayama S et al (2005) The transcriptional landscape of the mammalian genome. Science (80-) 309(5740):1559 LP – 1563. https://doi.org/10.1126/science.1112014
Guttman M, Amit I, Garber M et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235):223–227. https://doi.org/10.1038/nature07672
Iyer MK, Niknafs YS, Malik R et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47(3):199–208. https://doi.org/10.1038/ng.3192
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43(6):904–914. https://doi.org/10.1016/j.molcel.2011.08.018
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10(6):924–933. https://doi.org/10.4161/rna.24604
Bhan A, Soleimani M, Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res 77(15):3965 LP – 3981. https://doi.org/10.1158/0008-5472.CAN-16-2634
Li Z, Xu C, Ding B, Gao M, Wei X, Ji N (2017) Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J Neurooncol 134(1):19–28. https://doi.org/10.1007/s11060-017-2498-5
Liu Q, Yu W, Zhu S et al (2019) Long noncoding RNA GAS5 regulates the proliferation, migration, and invasion of glioma cells by negatively regulating miR-18a-5p. J Cell Physiol 234(1):757–768. https://doi.org/10.1002/jcp.26889
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. https://doi.org/10.1146/annurev-biochem-051410-092902
Khalil AM, Guttman M, Huarte M et al (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci. 106(28):11667 LP – 11672. https://doi.org/10.1073/pnas.0904715106
Schmitt AM, Chang HY (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29(4):452–463. https://doi.org/10.1016/j.ccell.2016.03.010
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100(1):57–70. https://doi.org/10.1016/S0092-8674(00)81683-9
Li X, Meng Y (2019) Survival analysis of immune-related lncRNA in low-grade glioma. BMC Cancer 19(1):813. https://doi.org/10.1186/s12885-019-6032-3
Lu R, Chen J, Kong L, Zhu H (2018) Prognostic value of lncRNA ROR expression in various cancers: a meta-analysis. Biosci Rep. 38(5). https://doi.org/10.1042/BSR20181095
Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J (2018) An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol Neurobiol 55(5):3684–3697. https://doi.org/10.1007/s12035-017-0572-9
Shen H, Mo Q, Xu X, Liu B (2020) The prognostic value of lncRNA SNHG6 in cancer patients. Cancer Cell Int 20(1):286. https://doi.org/10.1186/s12935-020-01383-9
Raudvere U, Kolberg L, Kuzmin I et al (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47(W1):W191–W198. https://doi.org/10.1093/nar/gkz369
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6(7):e21800. https://doi.org/10.1371/journal.pone.0021800
Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J (2019) NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 47(W1):W234–W241. https://doi.org/10.1093/nar/gkz240
Fukunaga T, Iwakiri J, Ono Y, Hamada M (2019) LncRRIsearch: a web server for lncRNA-RNA interaction prediction integrated with tissue-specific expression and subcellular localization data . Front Genet 10:462. https://www.frontiersin.org/article/https://doi.org/10.3389/fgene.2019.00462.
Shannon Paul, Markiel Andrew, Ozier Owen, Baliga Nitin S., Wang Jonathan T., Ramage Daniel, Amin Nada, Schwikowski Benno, Ideker Trey (2003) Cytoscape: a Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Bader GD, Hogue CWV (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 4(1):2. https://doi.org/10.1186/1471-2105-4-2
Goedhart Joachim, uijsterburg MS (2020) VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci Rep 10(1):20560. https://doi.org/10.1038/s41598-020-76603-3
Lever J, Zhao EY, Grewa J, Jones MR, Jones SJM (2019) CancerMine: a literature-mined resource for drivers, oncogenes and tumor suppressors in cancer. Nat Methods 16(6):505–507. https://doi.org/10.1038/s41592-019-0422-y
Rajakulendran S, Roberts J, Koltzenburg M, Hanna MG, Stewart H (2013) Deletion of chromosome 12q21 affecting KCNC2 and ATXN7L3B in a family with neurodevelopmental delay and ataxia. J Neurol Neurosurg Psychiatry 84(11):1255 LP – 1257. https://doi.org/10.1136/jnnp-2012-304555
Ivanov SV, Panaccione A, Brown B et al (2013) TrkC signaling is activated in adenoid cystic carcinoma and requires NT-3 to stimulate invasive behavior. Oncogene 32(32):3698–3710. https://doi.org/10.1038/onc.2012.377
Lee MTM, Chen CH, Lee CS et al (2011) Genome-wide association study of bipolar I disorder in the Han Chinese population. Mol Psychiatry 16(5):548–556. https://doi.org/10.1038/mp.2010.43
Oliver EE, Hughes EK, Puckett MK, Chen R, Lowther WT, Howlett AC (2020) Cannabinoid receptor interacting protein 1a (CRIP1a) in health and disease. Biomolecules. 10(12). https://doi.org/10.3390/biom10121609
Cocco E, Scaltriti M, Drilon A (2018) NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 15(12):731–747. https://doi.org/10.1038/s41571-018-0113-0
Liu X, Lei Q, Yu Z, et al (2015) MiR-101 reverses the hypomethylation of the LMO3 promoter in glioma cells. Oncotarget; 6, No 10. https://www.oncotarget.com/article/3181/text/.
Yang JH, Tucker SM, Levy ML, Crawford JR (2021) Rare case of BRAF V600E mutant anaplastic pleomorphic xanthroastrocytoma in a 5-year survivor of acute lymphoblastic leukaemia. BMJ Case Rep 14(2):e241815. https://doi.org/10.1136/bcr-2021-241815
Suzuki H, Yoshida T, Morisada N et al (2019) De novo NSF mutations cause early infantile epileptic encephalopathy. Ann Clin Transl Neurol 6(11):2334–2339. https://doi.org/10.1002/acn3.50917
Lindström MS (2019)Expanding the scope of candidate prognostic marker IGFBP2 in glioblastoma. Biosci Rep. 39(7). https://doi.org/10.1042/BSR20190770
Gandini NA, Fermento ME, Salomón DG et al (2014) Heme oxygenase-1 expression in human gliomas and its correlation with poor prognosis in patients with astrocytoma. Tumor Biol 35(3):2803–2815. https://doi.org/10.1007/s13277-013-1373-z
Liang P, Chai Y, Zhao H, Wang G (2020) Predictive analyses of prognostic-related immune genes and immune infiltrates for glioblastoma. Diagnostics. 10(3). https://doi.org/10.3390/diagnostics10030177
Khan S, Lu X, Huang Q et al (2019) IGFBP2 plays an essential role in cognitive development during early life. Adv Sci 6(23):1901152. https://doi.org/10.1002/advs.201901152
Dunn LL, Kong SMY, Tumanov S et al (2021) Hmox1 (heme oxygenase-1) protects against ischemia-mediated injury via stabilization of HIF-1α (hypoxia-inducible factor-1α). Arterioscler Thromb Vasc Biol 41(1):317–330. https://doi.org/10.1161/ATVBAHA.120.315393
Li T, Forbes ME, Fuller GN, Li J, Yang X, Zhang W (2020) IGFBP2: integrative hub of developmental and oncogenic signaling network. Oncogene 39(11):2243–2257. https://doi.org/10.1038/s41388-020-1154-2
Hollingworth P, Harold D, Sims R et al (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43(5):429–435. https://doi.org/10.1038/ng.803
Irvine M, Stewart A, Pedersen B, Boyd S, Kefford R, Rizos H (2018) Oncogenic PI3K/AKT promotes the step-wise evolution of combination BRAF/MEK inhibitor resistance in melanoma. Oncogenesis 7(9):72. https://doi.org/10.1038/s41389-018-0081-3
Del Bufalo F, Ceglie G, Cacchione A, et al (2018) BRAF V600E inhibitor (vemurafenib) for BRAF V600E mutated low grade gliomas . Front Oncol 8:526. https://www.frontiersin.org/article/https://doi.org/10.3389/fonc.2018.00526.
Weber RG, Hoischen A, Ehrler M et al (2007) Frequent loss of chromosome 9, homozygous CDKN2A/p14ARF/CDKN2B deletion and low TSC1 mRNA expression in pleomorphic xanthoastrocytomas. Oncogene 26(7):1088–1097. https://doi.org/10.1038/sj.onc.1209851
Mistry M, Zhukova N, Merico D et al (2015) BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol 33(9):1015–1022. https://doi.org/10.1200/JCO.2014.58.3922
Yang L, Xue Y, Liu J et al (2017) Long noncoding RNA ASAP1-IT1 promotes cancer stemness and predicts a poor prognosis in patients with bladder cancer. Neoplasma 64(6):847–855. https://doi.org/10.4149/neo_2017_606
Guo L, Zhou Y, Chen Y, Sun H, Wang Y, Qu Y (2018) LncRNA ASAP1-IT1 positively modulates the development of cholangiocarcinoma via hedgehog signaling pathway. Biomed Pharmacother 103:167–173. https://doi.org/10.1016/j.biopha.2018.04.015
Wang L, Sun L, Liu R et al (2021) Long non-coding RNA MAPKAPK5-AS1/PLAGL2/HIF-1α signaling loop promotes hepatocellular carcinoma progression. J Exp Clin Cancer Res 40(1):72. https://doi.org/10.1186/s13046-021-01868-z
Dong P, Xiong Y, Yue J, et al (2018) Long non-coding RNA NEAT1: a novel target for diagnosis and therapy in human tumors. Front Genet 9:471. https://www.frontiersin.org/article/https://doi.org/10.3389/fgene.2018.00471.
Ahmadov U, Picard D, Bartl J et al (2021) The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness and radiotherapy resistance in glioblastoma. Cell Death Dis 12(10):885. https://doi.org/10.1038/s41419-021-04146-0
Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT (2009) GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28(2):195–208. https://doi.org/10.1038/onc.2008.373
Gao Q, Xie H, Zhan H, Li J, Liu Y, Huang W (2017) Prognostic values of long noncoding RNA GAS5 in various carcinomas: an updated systematic review and meta-analysis. Front Physiol 8:814. https://www.frontiersin.org/article/https://doi.org/10.3389/fphys.2017.00814.
Cummings J, Ethell BT, Jardine L, et al (2003) Glucuronidation as a mechanism of intrinsic drug resistance in human colon cancer. Cancer Res. 63(23):8443 LP - 8450. http://cancerres.aacrjournals.org/content/63/23/8443.abstract.
Garvin AJ, Morris JR (2017) SUMO, a small, but powerful, regulator of double-strand break repair. Philos Trans R Soc B Biol Sci 372(1731):20160281. https://doi.org/10.1098/rstb.2016.0281
Liu S, Liang J, Liu Z et al (2021) The role of CD276 in cancers. Front Oncol 11. https://www.frontiersin.org/article/10.3389/fonc.2021.654684
Liu Y, Yang Y, Zhang L et al (2021) LncRNA ASAP1-IT1 enhances cancer cell stemness via regulating miR-509-3p/YAP1 axis in NSCLC. Cancer Cell Int 21(1):572. https://doi.org/10.1186/s12935-021-02270-7
Deng L, Meng T, Chen L, Wei W, Wang P (2020) The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther 5(1):11. https://doi.org/10.1038/s41392-020-0107-0
Fang X, Zhou W, Wu Q et al (2016) Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. J Exp Med 214(1):245–267. https://doi.org/10.1084/jem.20151673
Murphy JM, Rodriguez YAR, Jeong K, Ahn E-YE, Lim S-TS (2020) Targeting focal adhesion kinase in cancer cells and the tumor microenvironment. Exp Mol Med. 52(6):877–886. https://doi.org/10.1038/s12276-020-0447-4
Chen Z, Borek D, Padrick SB et al (2010) Structure and control of the actin regulatory WAVE complex. Nature 468(7323):533–538. https://doi.org/10.1038/nature09623
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D (2019) LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 47(D1):D1034–D1037. https://doi.org/10.1093/nar/gky905
Funding
We are thankful for financial support for funds and support received from various funding agencies. R. K. is thankful to IIT Delhi internal grant. C. S. gratefully acknowledges the funding support received from the J C Bose Fellowship of the Department of Science and Technology (DST). I. D. is thankful to the Department of Biotechnology (DBT) for the Junior Research Fellowship. R. G. is thankful to IIT Delhi for Senior Research Fellowship. J. S. is thankful to the Indian Council of Medical Research (ICMR) for the Senior Research Fellowship.
Author information
Authors and Affiliations
Contributions
C. S., R. K., and V. S. conceived this project and supervised the study. A. S. performed surgery and provided the tumor sections. P. J., J. S., I. D., C. S., M. C. S., and V. S. performed the histological analyses and harvested RNA for gene expression profiling. N. S. performed pre-processing of the microarray data. R. K., R. G., I. D., and J. S. analyzed and interpreted the gene expression data and were the major contributors to writing the manuscript. All authors were involved in the editing of the manuscript. R. G. and R. K. analyzed the lncRNA and lncRNA-mRNA co-expression data, and performed pathway and network analyses. V. S. and R.G. plotted the Kaplan–Meier curves. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
Ethics approval was taken from AIIMS Ethics Institute (IEC-724, 04/10/2019).
Consent to Participate
Yes.
Consent for Publication
Yes.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12035_2022_2893_MOESM1_ESM.xlsx
Supplementary file1 TableS1: A list of differentially regulated mRNAs in PXA versus control brain,obtained by applying log2 fold change cut-off. (XLSX 1154 KB)
12035_2022_2893_MOESM2_ESM.xlsx
Supplementary file2 TableS2: A list of differentially regulated lncRNAs in PXA versus control brain,obtained by applying log2 fold change cut-off. (XLSX 157 KB)
12035_2022_2893_MOESM3_ESM.ps
Supplementary file3 Fig.1: A depiction of non-redundant GO BP and GO MF terms of the top 100 most differentially expressed mRNAs in PXA using http://revigo.irb.hr/ online tool. a GO BP of upregulated mRNAs. b GO BPof downregulated mRNAs. c GO MF of upregulated mRNAs. d GO MF of downregulatedmRNAs (PS 575 KB)
12035_2022_2893_MOESM4_ESM.xlsx
Supplementary file4 TableS3: A list of the 100 most deregulated lncRNAs’ predicted transcripts obtainedby arranging as per ‘Sum of energy’ values using ‘LncRRiSearch’ webtoolhttp://rtools.cbrc.jp/LncRRIsearch/. (XLSX 174 KB)
12035_2022_2893_MOESM5_ESM.ps
Supplementary file5 Fig.2: A depiction of non-redundant GO BP and GO MF terms of the top 100 most differentially expressed lncRNAs in PXA using http://revigo.irb.hr/ onlinetool. a GO MF of upregulated lncRNAs. b GO MF of downregulated lncRNAs. c GO BPof upregulated lncRNAs. d GO BP of downregulated lncRNAs. (PS 514 KB)
12035_2022_2893_MOESM6_ESM.xlsx
Supplementary file6 TableS4: A list of pathway terms reported from the lncRNA target PPI network using‘g:Profiler’ online tool https://biit.cs.ut.ee/gprofiler/. (XLSX 21 KB)
12035_2022_2893_MOESM7_ESM.xlsx
Supplementary file7 Table S5: Differentially expressed mRNAs in PXA as predicted targets of the differentially expressed lncRNAs using‘LncRRiSearch’ webtool http://rtools.cbrc.jp/LncRRIsearch/. List made byintersecting the predicted targets of lncRNAs with the list of differentially regulated genes in PXA. (XLSX 22 KB)
12035_2022_2893_MOESM8_ESM.xlsx
Supplementary file8 TableS6: Expression correlation output file as obtained from the ‘Expression correlation’ plugin of Cytoscape by choosing >0.85 as the Pearson’scorrelation cut-off (r). (XLSX 778 KB)
12035_2022_2893_MOESM9_ESM.xlsx
Supplementary file9 TableS7: A list of pathways for the top 30 co-expressed and differentially regulated lncRNA-mRNA pairs obtained using ‘g:Profiler’ online toolhttps://biit.cs.ut.ee/gprofiler/. (XLSX 10 KB)
12035_2022_2893_MOESM10_ESM.xlsx
Supplementary file10 TableS8: Progression free survival data input file for the 20 most differentially regulated lncRNAs in PXA. Median expression threshold set for dividing thepatients in low and high expression groups. (XLSX 41 KB)
12035_2022_2893_MOESM11_ESM.xlsx
Supplementary file11 TableS9: Progression free survival data input file for the 20 most differentially regulated mRNAs in PXA. Median expression threshold set for dividing thepatients in low and high expression groups. (XLSX 199 KB)
Rights and permissions
About this article
Cite this article
Dandapath, I., Gupta, R., Singh, J. et al. Long Non-coding RNA and mRNA Co-expression Network Reveals Novel Players in Pleomorphic Xanthoastrocytoma. Mol Neurobiol 59, 5149–5167 (2022). https://doi.org/10.1007/s12035-022-02893-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02893-5