Abstract
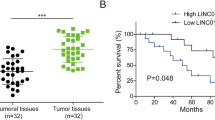
The newly discovered LINC02532 is abnormally expressed in a variety of cancers and promotes cancer progression. The research proposed to discover the biological and molecular mechanisms of LINC02532 in breast cancer (BCa). In the resected BCa tissue samples and adjacent normal tissues, LINC02532, miR-541-3p, and High Mobility Group A1 (HMGA1) levels were determined. Cell function experiments were carried out on the premise of cell transfection with relevant plasmids. Based on that, the influence of LINC02532, miR-541-3p, and HMGA1 on MCF-7 cell activities (proliferation, migration, invasion, cell cycle, and apoptosis) was determined, as well as on EMT. Additionally, animal experiments were allowed to support cell experimental conclusions on LINC02532. Finally, the mechanistic network of LINC02532, miR-541-3p, and HMGA1 was identified. It was BCa tissues highly expressing LINC02532 and HMGA1, while lowly expressing miR-541-3p. Functionally, LINC02532 depletion repressed the activities and EMT process of MCF-7 cells. Silencing LINC02532 delayed tumor growth in mice. In terms of mechanism, LINC02532 mainly existed in the cytoplasm and could mediate HMGA1 expression by absorbing miR-541-3p. The findings offer new insights into the molecular mechanisms of LINC02532 in BCa and, more importantly, new strategies for the clinical treatment of BCa.







Similar content being viewed by others
Data Availability
Data is available from the corresponding author on request.
References
Bray, F., et al. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. C Ca: A Cancer Journal for Clinicians, 68(6), 394–424.
Dabi, Y., et al. (2022). Overview of non-coding RNAs in breast cancers. Transl Oncol, 25, 101512.
Siegel, R. L., Miller, K. D., & Jemal, A. (2020). Cancer statistics, 2020. C Ca: A Cancer Journal for Clinicians, 70(1), 7–30.
Gibb, E. A., Brown, C. J., & Lam, W. L. (2011). The functional role of long non-coding RNA in human carcinomas. Molecular Cancer, 10, 38.
Yao, R. W., Wang, Y., & Chen, L. L. (2019). Cellular functions of long noncoding RNAs. Nature Cell Biology, 21(5), 542–551.
Kondo, Y., Shinjo, K., & Katsushima, K. (2017). Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Science, 108(10), 1927–1933.
Hanniford, D., et al. (2020). Epigenetic silencing of CDR1as drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell, 37(1), 55–70e15.
Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nature Medicine, 21(11), 1253–1261.
Huo, X., et al. (2017). Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: Implications for tumorigenesis, Disease progression, and Liver cancer stem cells. Molecular Cancer, 16(1), 165.
Flores-Huerta, N. (2021). LncRNAs and microRNAs as essential regulators of stemness in Breast Cancer stem cells. Biomolecules, 11(3).
Yang, Y. X., et al. (2018). Long non-coding RNA p10247, high expressed in Breast cancer (lncRNA-BCHE), is correlated with Metastasis. Clinical & Experimental Metastasis, 35(3), 109–121.
Gupta, R. A., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer Metastasis. Nature, 464(7291), 1071–1076.
Lakshmi, S., Hughes, T. A., & Priya, S. (2021). Exosomes and exosomal RNAs in Breast cancer: A status update. European Journal of Cancer, 144, 252–268.
Bliss, S. A., et al. (2016). Mesenchymal stem cell-derived Exosomes Stimulate Cycling quiescence and early Breast Cancer Dormancy in Bone Marrow. Cancer Research, 76(19), 5832–5844.
Zhang, C., et al. (2019). Novel long non-coding RNA LINC02532 promotes gastric cancer cell proliferation, migration, and invasion in vitro. World Journal of Gastrointestinal Oncology, 11(2), 91–101.
Zhou, X. (2021). LINC02532 contributes to Radiosensitivity in Clear Cell Renal Cell Carcinoma through the miR-654-5p/YY1 Axis. Molecules, 26(22).
Chen, D., et al. (2018). MicroRNA-129-5p regulates glycolysis and cell proliferation by targeting the glucose transporter SLC2A3 in gastric Cancer cells. Frontiers in Pharmacology, 9, 502.
Li, J. (2021). MiR-490-5p Restrains Progression of Gastric cancer through DTL Repression Gastroenterol Res Pract, 2021: p. 2894117.
Wu, D., et al. (2021). STAT3-induced HLA-F-AS1 promotes cell proliferation and stemness characteristics in triple negative Breast cancer cells by upregulating TRABD. Bioorganic Chemistry, 109, 104722.
Xu, Q., et al. (2020). CircIL4R facilitates the tumorigenesis and inhibits ferroptosis in hepatocellular carcinoma by regulating the miR-541-3p/GPX4 axis. Cell Biology International, 44(11), 2344–2356.
Long, B., et al. (2018). Long noncoding RNA LOXL1-AS1 regulates Prostate cancer cell proliferation and cell cycle progression through mir-541-3p and CCND1. Biochemical and Biophysical Research Communications, 505(2), 561–568.
He, Z., et al. (2021). Mir-541-3p enhances the radiosensitivity of Prostate cancer cells by inhibiting HSP27 expression and downregulating beta-catenin. Cell Death Discov, 7(1), 18.
Sgarra, R., et al. (2018). High mobility Group A (HMGA) proteins: Molecular instigators of Breast cancer onset and progression. Biochimica Et Biophysica Acta - Reviews on Cancer, 1869(2), 216–229.
Reeves, R. (2010). Nuclear functions of the HMG proteins. Biochimica Et Biophysica Acta, 1799(1–2), 3–14.
Sgarra, R., et al. (2004). Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. Febs Letters, 574(1–3), 1–8.
Pegoraro, S., et al. (2013). HMGA1 promotes metastatic processes in basal-like Breast cancer regulating EMT and stemness. Oncotarget, 4(8), 1293–1308.
Shah, S. N., et al. (2013). HMGA1: A master regulator of Tumor progression in triple-negative Breast cancer cells. PLoS One, 8(5), e63419.
Pegoraro, S., et al. (2015). A novel HMGA1-CCNE2-YAP axis regulates Breast cancer aggressiveness. Oncotarget, 6(22), 19087–19101.
Dowsett, M., et al. (2011). Assessment of Ki67 in Breast cancer: Recommendations from the International Ki67 in Breast Cancer working group. Journal of the National Cancer Institute, 103(22), 1656–1664.
Zabaglo, L., et al. (2010). Comparative validation of the SP6 antibody to Ki67 in Breast cancer. Journal of Clinical Pathology, 63(9), 800–804.
Cuzick, J., et al. (2011). Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the genomic health recurrence score in early Breast cancer. Journal of Clinical Oncology, 29(32), 4273–4278.
Bartha, A., & Gyorffy, B. (2021). TNMplot.com: A web Tool for the comparison of Gene expression in normal, Tumor and metastatic tissues. International Journal of Molecular Sciences, 22(5).
Zhou, X. (Nov. 2021). LINC02532 Contributes to Radiosensitivity in Clear Cell Renal Cell Carcinoma through the miR-654-5p/YY1 Axis. Molecules (Basel, Switzerland) vol. 26,22 7040. 22.
Wu, Z., et al. (2020). Long noncoding RNA LINC00963 promotes Breast cancer progression by functioning as a molecular sponge for microRNA-625 and thereby upregulating HMGA1. Cell Cycle, 19(5), 610–624.
Cech, T. R., & Steitz, J. A. (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell, 157(1), 77–94.
Huang, Q. Y. (2019). Long non-coding RNA: Dual effects on Breast Cancer Metastasis and clinical applications. Cancers (Basel), 11(11).
Han, C., et al. (2019). CCAT1 promotes triple-negative Breast cancer progression by suppressing miR-218/ZFX signaling. Aging (Albany NY), 11(14), 4858–4875.
Tu, Z., et al. (2019). Microenvironmental Regulation of Long Noncoding RNA LINC01133 promotes Cancer Stem Cell-Like phenotypic traits in Triple-negative breast cancers. Stem Cells, 37(10), 1281–1292.
Vidovic, D., et al. (2020). ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death and Differentiation, 27(1), 363–378.
Kong, X., et al. (2019). LncRNA-CDC6 promotes Breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. Journal of Cellular Physiology, 234(6), 9105–9117.
Ye, G., et al. (2022). circ_0041732 promotes Breast Cancer Progression. Molecular Cancer Research, 20(10), 1561–1573.
Hu, C. Y., et al. (2021). Long non-coding RNA NORAD promotes the Prostate cancer cell extracellular vesicle release via microRNA-541-3p-regulated PKM2 to induce bone Metastasis of Prostate cancer. Journal of Experimental & Clinical Cancer Research : Cr, 40(1), 98.
Xia, Y. H., et al. (2019). Role of miR-541-3p/TMPRSS4 in the Metastasis and EMT of hepatocellular carcinoma. European Review for Medical and Pharmacological Sciences, 23(24), 10721–10728.
Lu, Y. J., et al. (2016). MiR-541-3p reverses cancer progression by directly targeting TGIF2 in non-small cell Lung cancer. Tumour Biology, 37(9), 12685–12695.
Leivonen, S. K., et al. (2014). High-throughput screens identify microRNAs essential for HER2 positive Breast cancer cell growth. Molecular Oncology, 8(1), 93–104.
Zanin, R., et al. (2019). HMGA1 promotes Breast cancer angiogenesis supporting the stability, nuclear localization and transcriptional activity of FOXM1. Journal of Experimental & Clinical Cancer Research : Cr, 38(1), 313.
Wang, L., et al. (2022). High mobility Group A1 (HMGA1): Structure, biological function, and therapeutic potential. International Journal of Biological Sciences, 18(11), 4414–4431.
Penzo, C. (2019). HMGA1 modulates gene transcription sustaining a Tumor Signalling pathway acting on the Epigenetic Status of Triple-negative Breast Cancer cells. Cancers (Basel), 11(8).
Mendez, O. (2019). Clinical implications of Extracellular HMGA1 in Breast Cancer. International Journal of Molecular Sciences, 20(23).
Treff, N. R., et al. (2004). High-mobility group A1a protein regulates Ras/ERK signaling in MCF-7 human Breast cancer cells. Oncogene, 23(3), 777–785.
Mansoori, B., et al. (2021). The synergy between mir-486-5p and tamoxifen causes profound cell death of tamoxifen-resistant Breast cancer cells. Biomedicine & Pharmacotherapy, 141, 111925.
Peluso, S. (2010). and Gennaro C. High-Mobility Group A (HMGA) Proteins and Breast Cancer Breast care (Basel, Switzerland) vol. 5,2 : 81–85.
Acknowledgements
Not applicable.
Funding
1. Shandong Provincial Natural Foundation (NO. ZR2020MH198). 2. Wu Jieping Medical Foundation Clinical Research Special Funding Fund (320.6750.2021-10-43).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interests
Authors declared no conflict of interest.
Ethical Approval
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All subjects was approved by Shandong Provincial Hospital Affiliated to Shandong First Medical University. And The animal experiment research protocol was approved by the Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong First Medical University and performed in accordance with the “Guidelines for the care and use of experimental animals.”
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
12033_2023_995_MOESM1_ESM.tif
Supplementary Material 1: Fig. 1 Upregulation of LINC02532 promotes proliferation, migration, and invasion of MDA-MB-231 cells, and inhibits cell apoptosis pcDNA3.1-LINC02532 was transfected into MDA-MB-231 cells. RT-qPCR detection of the transfection of pcDNA3.1-LINC02532 (A), analysis results of proliferation (B-C), migration and invasion (D), apoptosis rate (E), and cell cycle (F). Data were expressed as mean ± SD, * P < 0.05
12033_2023_995_MOESM2_ESM.tif
Supplementary Material 2: Fig. 2 LINC02532 promotes proliferation, migration, and invasion and inhibits apoptosis of MDA-MB-231 cells by regulating miR-654-5p. si-LINC02532-1 and miR-654-5p inhibitor were co-transfected into MDA-MB-231 cells, RT-qPCR detection of the transfection of si-LINC02532-1 and miR-654-5p inhibitor (A), analysis results of proliferation (B-C), migration and invasion (D), apoptosis rate (E) and cell cycle (F). Data were expressed as mean ± SD, * P < 0.05
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, C., Li, X., Pan, X. et al. LINC02532 by Mediating miR-541-3p/HMGA1 Axis Exerts a Tumor Promoter in Breast cancer. Mol Biotechnol (2023). https://doi.org/10.1007/s12033-023-00995-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-023-00995-6