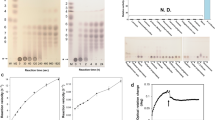
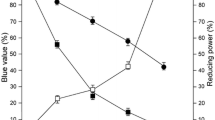
Abstract
Oenococcus oeni is the main bacterial species that drives malolactic fermentation in wine. Most O. oeni strains produce capsular exopolysaccharides (EPS) that may contribute to protect them in the wine hostile environment. In O. oeni genome sequences, several genes are predicted to encode priming glycosyltransferases (pGTs). These enzymes are essential for EPS formation as they catalyze the first biosynthetic step through the formation of a phosphoanhydride bond between a hexose-1-phosphate and a lipid carrier undecaprenyl phosphate. In many microorganisms, mutations abolishing the pGT activity also abolish the EPS formation. We first made an in silico analysis of all the genes encoding putative pGT over 50 distinct O. oeni genome sequences. Two polyisoprenyl-phosphate-hexose-1-phosphate transferases, WoaA and WobA, and a glycosyltransferase (It3) were particularly examined for their topology and amino acid sequence. Several isoforms of these enzymes were then expressed in E. coli, and their substrate specificity was examined in vitro. The substrate specificity varied depending on the protein isoform examined, and several mutations were shown to abolish WobA activity but not EPS synthesis. Further analysis of woaA and wobA gene expression levels suggests that WoaA could replace the deficient WobA and maintain EPS formation.




Similar content being viewed by others
Abbreviations
- EPS:
-
Exopolysaccharide
- eps :
-
Exopolysaccharide gene cluster
- pGT:
-
Priming glycosyltransferase
- it3:
-
Isolated gene of glycosyltransferase annotated as encoding a pGT
- WoaA:
-
pGT encoded in eps1 gene cluster
- WobA:
-
pGT encoded in eps2 gene cluster
References
Lonvaud-Funel, A. (1999). Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie van Leeuwenhoek, 76, 317–331.
Dols-Lafargue, M., Gindreau, E., Le Marrec, C., Chambat, G., Heyraud, A., & Lonvaud-Funel, A. (2007). Changes in red wine soluble polysaccharide composition induced by malolactic fermentation. Journal of Agriculture and Food Chemistry, 55, 9592–9599.
Dols-Lafargue, M., Lee, H. Y., Le Marrec, C., Heyraud, A., Chambat, G., & Lonvaud-Funel, A. (2008). Characterization of gtf, a glucosyltransferase gene in the genomes of Pediococcus parvulus and Oenococcus oeni, two bacterial species commonly found in wine. Applied and Environment Microbiology, 74, 4079–4090.
Dols-Lafargue, M., & Lonvaud-Funel, A. (2009). Polysaccharide production by grapes, must and wine microorganisms. In H. König, G. Uden, & J. Fröhlich (Eds.), Biology of microorganisms on grapes, in must and wine (pp. 243–260). Berlin, Heidelberg: Springer.
Dimopoulou, M., Hazo, L., & Dols-Lafargue, M. (2012). Exploration of phenomena contributing to the diversity of Oenococcus oeni exopolysaccharides. International Journal of Food Microbiology, 153, 114–122.
Ciezak, G., Hazo, L., Chambat, G., Heyraud, A., Lonvaud-Funel, A., & Dols-Lafargue, M. (2009). Evidence for exopolysaccharide production by Oenococcus oeni strains isolated from non-ropy wines. Journal of Applied Microbiology, 108, 499–509.
Dimopoulou, M., Vuillemin, M., Campbell-Sills, H., Lucas, P. M., Ballestra, P., Miot-Sertier, C., et al. (2014). Exopolysaccharide (EPS) synthesis by Oenococcus oeni: From genes to phenotypes. PLoS ONE, 9, e98898.
Dimopoulou, M., Bardeau, T., Ramonet, P. Y., Miot-Sertier, C., Claisse, O., Doco, T., et al. (2016). Exopolysaccharides produced by Oenococcus oeni: From genomic and phenotypic analysis to technological valorization. Food Microbiology, 53, 10–17.
Yother, J. (2011). Capsules of Streptococcus pneumoniae and other bacteria: Paradigms for polysaccharide biosynthesis and regulation. Annual Review of Microbiology, 65, 563–581.
Whitfield, C. (2006). Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annual Review of Biochemistry, 75, 39–68.
Ullrich, M. (2009). Bacterial polysaccharides: Current innovations and future trends. Poole: Horizon Scientific Press.
Valvano, M. A. (2003). Export of O-specific lipopolysaccharide. Frontiers in Bioscience, 8, s452–s471.
Wang, L., Liu, D., & Reeves, P. R. (1996). C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. Journal of Bacteriology, 178, 2598–2604.
James, D. B. A., Gupta, K., Hauser, J. R., & Yother, J. (2013). Biochemical activities of Streptococcus pneumoniae serotype 2 capsular glycosyltransferases and significance of suppressor mutations affecting the initiating glycosyltransferase Cps2E. Journal of Bacteriology, 195, 5469–5478.
Shainheit, M. G., Valentino, M. D., Gilmore, M. S., & Camilli, A. (2015). Mutations in pneumococcal cpsE generated via in vitro serial passaging reveal a potential mechanism of reduced encapsulation utilized by a conjunctival isolate. Journal of Bacteriology, 197, 1781–1791.
Kolkman, M. A., Morrison, D. A., Van Der Zeijst, B. A., & Nuijten, P. J. (1996). The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: Identification of the glycosyl transferase gene cps14E. Journal of Bacteriology, 178, 3736–3741.
van Kranenburg, R., Vos, H. R., van Swam, I. I., Kleerebezem, M., & de Vos, W. M. (1999). Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: Complementation, expression, and diversity. Journal of Bacteriology, 181, 6347–6353.
Dabour, N., & LaPointe, G. (2005). Identification and molecular characterization of the chromosomal exopolysaccharide biosynthesis gene cluster from Lactococcus lactis subsp. cremoris SMQ-461. Applied and Environment Microbiology, 71, 7414–7425.
Lebeer, S., Verhoeven, T. L., Francius, G., Schoofs, G., Lambrichts, I., Dufrêne, Y., et al. (2009). Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Applied and Environment Microbiology, 75, 3554–3563.
Remus, D. M., van Kranenburg, R., van Swam, I. I., Taverne, N., Bongers, R. S., Wels, M., et al. (2012). Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microbial Cell Factories, 11, 149.
Horn, N., Wegmann, U., Dertli, E., Mulholland, F., Collins, S. R. A., Waldron, K. W., et al. (2013). Spontaneous mutation reveals influence of exopolysaccharide on Lactobacillus johnsonii surface characteristics. PLoS ONE, 8, e59957.
Schaffner, T. O., Hinds, J., Gould, K. A., Wüthrich, D., Bruggmann, R., Küffer, M., et al. (2014). A point mutation in cpsE renders Streptococcus pneumoniae nonencapsulated and enhances its growth, adherence and competence. BMC Microbiology, 14, 210.
Goudenège, D., Boursicot, V., Versigny, T., Bonnetot, S., Ratiskol, J., Sinquin, C., et al. (2014). Genome sequence of Vibrio diabolicus and identification of the exopolysaccharide HE800 biosynthesis locus. Applied Microbiology and Biotechnology, 98, 10165–10176.
De Man, J. C., Rogosa, M., & Sharpe, M. E. (1960). A medium for the cultivation of lactobacilli. Journal of Applied Bacteriology, 23, 130–135.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Kolkman, M. A. B., Wakarchuk, W., Nuijten, P. J. M., & Van Der Zeijst, B. A. M. (1997). Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: Molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Molecular Microbiology, 26, 197–208.
Stingele, F., Newell, J. W., & Neeser, J. R. (1999). Unraveling the function of glycosyltransferases in Streptococcus thermophilus Sfi6. Journal of Bacteriology, 181, 6354–6360.
Desroches, N., Beltramo, C., & Guzzo, J. (2005). Determination of an internal control to apply reverse transcription quantitative PCR to study stress response in the lactic acid bacterium Oenococcus oeni. Journal of Microbiological Methods, 60, 325–333.
Cartee, R. T., Forsee, W. T., Bender, M. H., Ambrose, K. D., & Yother, J. (2005). CpsE from type 2 Streptococcus pneumoniae catalyzes the reversible addition of glucose-1-phosphate to a polyprenyl phosphate acceptor, initiating type 2 capsule repeat unit formation. Journal of Bacteriology, 187, 7425–7433.
Patel, K. B., Ciepichal, E., Swiezewska, E., & Valvano, M. A. (2012). The C-terminal domain of the Salmonella enterica WbaP (UDP-galactose:Und-P galactose-1-phosphate transferase) is sufficient for catalytic activity and specificity for undecaprenyl monophosphate. Glycobiology, 22, 116–122.
Ardiccioni, C., Clarke, O. B., Tomasek, D., Issa, H. A., von Alpen, D. C., Pond, H. L., et al. (2016). Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis. Nature Communications, 7, 10175.
Guan, S., Bastin, D. A., & Verma, N. K. (1999). Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology, 145, 1263–1273.
Stingele, F., Neeser, J. R., & Mollet, B. (1996). Identification and characterization of the eps (Exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. Journal of Bacteriology, 178, 1680–1690.
Minic, Z., Marie, C., Delorme, C., Faurie, J. M., Mercier, G., Ehrlich, D., et al. (2007). Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase. Journal of Bacteriology, 189, 1351–1357.
Steiner, K., Novotny, R., Patel, K., Vinogradov, E., Whitfield, C., Valvano, M. A., et al. (2007). Functional characterization of the initiation enzyme of S-layer glycoprotein glycan biosynthesis in Geobacillus stearothermophilus NRS 2004/3a. Journal of Bacteriology, 189, 2590–2598.
Qi, X., Ma, M., Wang, L., Zhang, Y., Jiang, R., Bai, L., et al. (2015). Biochemical characterization of a novel bifunctional glycosyl-1-phosphate transferase involved in the exopolysaccharide biosynthesis. Biochemical and Biophysical Research Communications, 465, 113–118.
Pelosi, L., Boumedienne, M., Saksouk, N., Geiselmann, J., & Geremia, R. A. (2005). The glucosyl-1-phosphate transferase WchA (Cap8E) primes the capsular polysaccharide repeat unit biosynthesis of Streptococcus pneumoniae serotype 8. Biochemical and Biophysical Research Communications, 327, 857–865.
Xayarath, B., & Yother, J. (2007). Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. Journal of Bacteriology, 189, 3369–3381.
Yasuda, E., Serata, M., & Sako, T. (2008). Suppressive effect on activation of macrophages by Lactobacillus casei strain Shirota genes determining the synthesis of cell wall-associated polysaccharides. Applied and Environment Microbiology, 74, 4746–4755.
Lee, I. C., Caggianiello, G., van Swam, I. I., Taverne, N., Meijerink, M., Bron, P. A., et al. (2016). Strain-specific features of extracellular Polysaccharides and their impact on Lactobacillus plantarum-host interactions. Applied and Environment Microbiology, 82(13), 3959–3970.
Campbell-Sills, H., El Khoury, M., Favier, M., Romano, A., Biasioli, F., Spano, G., et al. (2015). Phylogenomic analysis of Oenococcus oeni reveals specific domestication of strains to cider and wines. Genome Biology and Evolution, 7, 1506–1518.
Sternes, P. R., & Borneman, A. R. (2016). Consensus pan-genome assembly of the specialized wine bacterium Oenococcus oeni. BMC Genomics, 17, 308.
Acknowledgements
This work was supported by the French National research Agency [Grant Number ANR-10-ALIA-003].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors do not have potential conflict of interest.
Ethical Approval
The article does not include any study with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dimopoulou, M., Claisse, O., Dutilh, L. et al. Molecular Cloning, Expression and Characterization of Oenococcus oeni Priming Glycosyltransferases. Mol Biotechnol 59, 323–333 (2017). https://doi.org/10.1007/s12033-017-0021-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-017-0021-z