Abstract
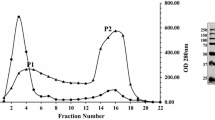
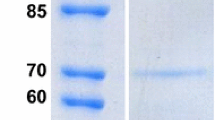
The filamentous fungus Sclerotinia sclerotiorum produces a complete set of cellulolytic enzymes needed for efficient solubilization of native cellulose, the major component of plants. In this work, we reported the molecular characterization of an important glycosyl-hydrolase enzyme classified as endo-β-1,4-glucanase. The importance of this enzyme was revealed with the in-gel activity staining, showing a high degradation capacity of cellulose. When purified from native gel and ran in denaturing polyacrylamide gel, the polypeptide has an apparent molecular mass of about 34 kDa called Endo2. For further characterization of this protein, a mass spectrometry approach was carried out. The LC–MS/MS analysis revealed two peptides belonging to this enzyme. The genomic DNA and cDNA sequences were resolved by PCR amplification and sequencing, revealing a gene with two intron sequences. The open reading frame of 987 bp encoded a putative polypeptide of 328 amino acids having a calculated molecular mass of 33,297 Da. Yet, the molecular modeling and comparative investigation of different 3D cellulase structures showed that this endoglucanase isoform has probably two domains. A core domain having a high similarity with endoglucanases family 5 and a cellulose-binding domain having similarities with those of exo-type cellulases of family 1, linked together by a serine-threonine-rich region. These results are with great interests and show new characteristics of S. sclerotiorum glucanase.






Similar content being viewed by others
References
Bhat, M. K., & Bhat, S. (1997). Cellulose degrading enzymes and their potential industrial applications. Biotechnology Advances, 15, 583–620.
Clarke, A. J. (1997). Biodegradation of Cellulose: Enzymology and Biotechnology. Lancaster, PA: Technomic Publishing Company Inc.
Ghosh, B. K., & Gosh, A. (1992). Degradation of cellulose by fungal cellulase. In G. Winkelmann (Ed.), Microbial degradation of natural products (pp. 84–116). New York: VCH Publisher Inc.
Beguin, P., & Aubert, J. P. (1994). The biological degradation of cellulose. FEMS Microbiology Reviews, 13, 25–58.
Lynd, L. R., Paul, J., Weimer, P. J., van Zyl, W. H., & Pretorius, I. S. (2002). Microbial cellulose utilization: Fundamentals and biotechnology. Microbiology and Molecular Biology Reviews, 66, 506–577.
Dienes, D., Egyhazi, A., & Reczey, K. (2004). Treatment of recycled fiber with Trichoderma cellulases. Industrial Crops and Products, 20, 11–21.
Duan, X. Y., Liu, S. Y., Zhang, W. C., Zhang, Q. X., & Gao, P. J. (2004). Volumetric productivity improvement for endogulcanase of Trichoderma pseudokoingii S-38. Journal of Applied Microbiology, 96, 772–776.
Gilkes, N. R., Henrissat, B., Kilburn, D. G., Miller, R. C., & Warren, R. A. J. (1991). Domains in microbial beta-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiological Reviews, 55, 303–315.
Burton, J., Wood, S. G., Pedyczak, A., & Siemieon, I. Z. (1989). Conformational preferences of sequential fragments of the hinge region of human IgA1 immunoglobulin molecule: II. Biophysical Chemistry, 33, 39–45.
Bushuev, V. N., Gudkov, A. T., Liljas, A., & Sepetov, N. F. (1989). The flexible region of protein L12 from bacterial ribosomes studied by proton nuclear magnetic resonance. Journal of Biological Chemistry, 264, 4498–4505.
Parry, N. J., Beever, D. E., Owen, E., Nerinck, W., Claeyssens, M., Beeumen, J. V., et al. (2002). Biochemical characterization and mode of action of a thermostable endoglucanase purified from Thermoascus aurantiacus. Archives of Biochemistry and Biophysics, 404, 243–253.
Ooi, T., Shinmyo, A., Okada, H., Murao, S., Kawaguchi, T., & Arai, M. (1990). Complete nucleotide sequence of a gene coding for Aspergillus aculeatus cellulase (F1-CMCase). Nucleic Acids Research, 18, 58–84.
Sakamoto, S., Tamura, G., Ito, K., Ishikawa, T., Iwano, K., & Nishiya, N. (1995). Cloning and sequencing of cellulase cDNA from Aspergillus kawachii and its expression in Saccharomyces cerevisiae. Current Genetics, 27, 435–439.
Hong, J., Hisanori, T., Shunichi, A., Yamamtot, K., & Kumagai, H. (2001). Cloning of a gene encoding a highly stable endo-β-1, 4-glucanase from Aspergillus niger and its expression in yeast. Journal of Bioscience and Bioengineering, 92, 434–439.
Sheppard, P. O., Grant, F. J., Oort, P. J., Sprecher, C. A., Foster, D. C., Hagen, F. S., et al. (1994). The use of conserved cellulase family-specific sequences to clone cellulase homologue cDNAs from Fusarium oxysporum. Gene, 150, 163–167.
Shuyan, L., Xinyuan, D., Xuemei, L., & Peiji, G. (2006). A novel thermophilic endoglucanase from a mesophilic fungus Fusarium oxysporum. Chinese Science Bulletin, 51, 191–197.
Saloheimo, A., Henrissat, B., Hoffren, A. M., Teleman, O., & Penttila, M. (1994). A novel small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Molecular Microbiology, 13, 219–228.
Bhat, M. K. (2004). Cellulases and related enzymes in biotechnology. Biotechnology Advances, 18, 355–383.
Purdy, L. H. (1979). Sclerotinia sclerotiorum: history, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology, 69, 875–880.
Lumsden, R. D. (1979). Histology and physiology of pathogenesis in plant diseases caused by Sclerotinia species. Phytopathology, 69, 890–896.
Lumdsen, R. D. (1969). Sclerotinia sclerotiorum infection of bean and the production of cellulase. Phytopathology, 59, 653–657.
Montenecourt, B. S., & Eveleigh, D. E. (1985). Fungal carbohydrases: Amylases and cellulases. In J. N. Bennett & L. L. Lasure (Eds.), Gene manipulations in fungi (pp. 491–509). Orlando, FL: Academic Press.
Smaali, M. I., Gargouri, M., Limam, F., Fattouch, S., Maugard, T., Legoy, M. D., et al. (2003). Production, purification and biochemical characterization of two β-glucosidases from Sclerotinia sclerotiorum. Applied Biochemistry and Biotechnology, 111, 29–40.
Ellouze, O., Mejri, M., Smaali, M. I., Limam, F., & Marzouki, M. N. (2007). Induction, properties and application of xylanase activity from Sclerotinia sclerotiorum fungus. Journal of Food Biochemistry, 31, 96–107.
Gargouri, M., Smaali, M. I., Maugard, T., Legoy, M. D., & Marzouki, M. N. (2004). Fungus β-glycosidases: immobilization and use in alkyl-β-glycoside synthesis. Journal of Molecular Catalysis B, 29, 89–94.
Mandels, M., & Weber, J. (1969). The production of cellulases. Advances in Chemistry Series, 95, 391–414.
Miller, G. L. (1959). Use of dinitrosalicyclic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
Laemmli, U. K., Molbert, E., Showe, M., & Kelenberger, E. (1970). Form-determining function of genes required for the assembly of the head of bacteriophage T4. Journal of Molecular Biology, 49, 99–113.
Ellouz, C. S., Mechichi, T., Limam, F., & Marzouki, M. N. (2005). Purification and characterization of two low molecular weight endoglucanases produced by Penicillium occitanis mutant pol 6. Applied Biochemistry and Biotechnology, 125, 99–112.
Blum, H., Beier, H., & Gross, B. (1987). Improved silver staining of plant proteins RNA and DNA in polyacrylamide gels. Electrophoresis, 8, 93–99.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: A laboratory mammal. New York: Cold Spring Harbor Laboratory Press.
Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucl. Acids Res., 16(22), 10881–10890.
Gouet, P., Robert, X., & Courcelle, E. (2003). ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Research, 31, 3320–3323.
Nielsen, H., Engelbrecht, J., Brunak, S., & Von Heijne, G. (1997). A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. International Journal of Neural Systems, 8, 581–599.
Schwede, T., Kopp, J., Guex, N., & Peitsch, M. C. (2003). SWISS-MODEL: an automated protein homology-modelling server. Nucleic Acids Research, 31, 3381–3385.
DeLano, W. L. (2002). The PyMOL molecular graphics system. San Carlos, CA: DeLano scientific.
Kubicek, C. P., Messner, R., Gruber, F., Mach, R. L., & Kubicek, P. E. M. (1993). The Trichoderma reesei cellulase regulatory puzzle from the interior of a secretory fungus. Enyzme and Microbial Technology, 15, 90–99.
Tangnu, K. S., Blanch, H. W., & Wilke, C. R. (1981). Enhanced production of cellulase, hemicellulase, and beta-glucosidase by Trichoderma reesei (Rut C-30). Biotechnology and Bioengineering, 23, 1837–1849.
Ryu, D. D., & Mandels, M. (1980). Cellulases: biosynthesis and applications. Enyzme and Microbial Technology, 2, 91–102.
Manheshwari, R., Bharadwaj, G., & Bhat, M. K. (2000). Physiology and enzymes of thermophilic fungi. Microbiology and Molecular Biology Reviews, 64, 461–488.
Vieille, C., & Zeikus, G. J. (2001). Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostablility. Microbiology and Molecular Biology Reviews, 65, 1–43.
Matsumoto, K., Endo, Y., Tamiya, N., Kano, M., Miyauchi, K., & Abe, J. (1974). Studies on cellulase produced by the phytopathogens: purification and enzymatic properties of cellulase of Fusarium moniliforme. Journal of Biochemistry, 76, 563–572.
Bennett, J. W., & Lasure, L. L. (1991). Growth media. In J. W. Bennett & L. L. Law-e (Eds.), More Gene Manipulations in Fungi (pp. 441–458). San Diego, CA: Academic Press.
Bhat, K. M., Mc Crae, S. I., & Wood, T. M. (1989). The endo-(1-4)-β-D-glucanase system of Penicillium pinophilum cellulase: isolation, purification and characterisation of five major endoglucanase components. Carbohydrate Research, 190, 279–297.
Gilbert, H. J., & Hazlewood, G. P. (1993). Bacterial cellulases and xylanases. Journal of General Microbiology, 139, 187–194.
Coughlan, M. P., & Ljungdahl, L. G. (1988). Comparative biochemistry of fungal and bacterial cellulolytic enzyme systems. In J. P. Aubert, P. Beguin, & J. Millet (Eds.), Biochemistry and Genetics of Cellulose Degradation (pp. 11–30). London: Academic Press.
Bhat, K. M., Gaikwad, J. S., & Maheshwari, R. (1993). Purification and characterisation of an extracellular β-glucosidase from the thermophilic fungus Sporotrichum thermophile and its influence on cellulase activity. Journal of General Microbiology, 139, 2825–2832.
Divne, C., Ståhlberg, J., Reinikainen, T., Ruohonen, L., Pettersson, G., Knowles, J. K. C., et al. (1994). The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science, 265, 524–528.
Henrissat, B., Teeri, T. T., & Warren, R. A. J. (1998). A scheme for designating enzymes that hydrolyze the polysaccharides in the cell walls of plants. FEBS Letters, 425, 352–354.
Henrissat, B. (1998). Enzymatic cellulose degradation. Cellulose Communication, 5, 84–90.
Arnold, K., Bordoli, L., Kopp, J., & Schwede, T. (2006). The SWISS-MODEL Workspace: A web based environment for protein structure homology modeling. Bioinformatics, 22, 195–201.
Davies, G. J., Dodson, G., Moore, M. H., Tolley, S. P., Dauter, Z., Wilson, K. S., et al. (1993). Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 Å resolution. Acta Crystallographica, 52, 7–17.
Davies, G. J., Tollay, S. P., Henrissat, B., Hjort, C., & Schulein, M. (1995). Structures of oligosaccharide bound forms of the endoglucanase V from Humicola insolens at 1.9 Å resolution. Biochemistry, 34, 16210–16220.
Davies, G. J., Dadson, G. G., Hubbart, R. E., Tolley, S. P., Dauter, Z., Wilson, K. S., et al. (1996). Structure and function of endoglucanase V. Nature, 365, 362–364.
Kleywegt, G. J., Zou, J. Y., Divne, C., Davies, G. J., Sinning, I., Stahlberg, J., et al. (1997). The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 Å resolution, and a comparison with related enzymes. Journal of Molecular Biology, 272, 383–397.
Sandgren, M., Shaw, A., Ropp, T. H., Wu, S., Bott, R., Cameron, A. D., et al. (2000). The X-ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 Å resolution. Journal of Molecular Biology, 308, 295–310.
Srisodsuk, M., Rainikainen, T., Panttila, M., & Teeri, T. T. (1993). Role of the interdomain linker peptide of Trichoderma reesei cellobiohydrolase I in its interaction with crystalline cellulose. Journal of Biological Chemistry, 268, 20756–20761.
Acknowledgments
We thank Dr. Thierry JOUENNE, director of the Laboratory of Polymers, Biopolymers and Surfaces in Rouen for providing the access to make mass spectrometry analysis and helpful discussions. This work is entirely financed by the Laboratory of Protein Engineering and Bioactive Molecules (LIP-MB) in the National Institute of Applied Sciences and Technology of Tunis, University of Carthage. The Tunisian Ministry of High Education, Scientific Research and Technology is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12033_2013_9714_MOESM1_ESM.docx
Collected data for mass spectrometry analysis. Raw data files were processed using Proteome Discoverer 1.3 software (Thermo Scientific). Peak lists were searched using the MASCOT search engine (Matrix Science). (DOCX 1,426 kb)
12033_2013_9714_MOESM2_ESM.docx
cDNA and genomic DNA sequences of the Endo2 β-1,4-endoglucanase of S. sclerotiorum. Introns are shown with silver background. Arrows indicates primers localization. (DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Chahed, H., Ezzine, A., Mlouka, A.B. et al. Biochemical Characterization, Molecular Cloning, and Structural Modeling of an Interesting β-1,4-Glucanase from Sclerotinia Sclerotiorum . Mol Biotechnol 56, 340–350 (2014). https://doi.org/10.1007/s12033-013-9714-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-013-9714-0