Abstract
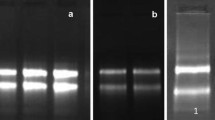
Jute (Corchorus capsularis), as a natural fibre producing plant species, ranks next to cotton only. Today, biotechnological approach has been considered as most accepted means for any genetic improvement of plant species. However, genetic control of the fibre development in jute has not yet been explored sufficiently for desired genetic improvement. One of the major impediments in exploring the genetic architecture in this crop at molecular level is the availability of good quality RNA from field-grown plant tissues mostly due to the presence of high amount of mucilage and phenolics. Development of a suitable RNA isolation method is becoming essential for deciphering developmental stage-specific gene expression pattern related to fibre formation in this crop species. A combination of modified hot borate buffer followed by isopycnic centrifugation (termed as HBIC) was adopted and found to be the best isolation method yielding sufficient quantity (~350–500 μg/gm fresh tissue) and good quality (A260/280 ratio 1.88 to 1.91) RNA depending on the developmental stage of stem tissue from field-grown jute plant. The poly A+ RNA purified from total RNA isolated by the present method was found amenable to efficient RT-PCR and cDNA library construction. The present development of RNA isolation was found to be appropriate for gene expression analysis related to fibre formation in this economically important jute plant in near future.







Similar content being viewed by others
Abbreviations
- PVPP:
-
Polyvinylpolypyrrolidone,
- DEPC:
-
Diethylpyrocarbonate
- CsCl:
-
Cesium chloride
- EDTA:
-
Ethylenediaminetetraacetic acid
- EtBr:
-
Ethidium bromide
References
Ahmed, S., Nabi, M. Z., Alam, M. M., Islam, M. S., Samira, R., Moosa, M. M., et al. (2009). A computational and experimental approach for developing jute ESTs from genomic clones. Australian Journal of Crop Science, 3(6), 322–328.
Alam, M. M., Sharmin, S., Nabi, Z., Mondal, S. I., Islam, M. S., Nayeem, S. B., et al. (2010). A putative leucine-rich repeat receptor-like kinase of jute involved in stress response. Plant Molecular Biology Reporter, 28(3), 394–402.
Bayer, C., Fay, M. F., & De Bruijn, A. Y. (1999). Support for an expanded family concept of Malvaceae within a recircumscribed order Malvales: a combined analysis of plastid atpB and rbcL DNA sequence. Botanical Journal of the Linnean Society, 129, 267–303.
Cathala, G., Savouret, J. F., Mendez, B., West, B. L., Karin, M., Martial, J. A., et al. (1983). A method for isolation of intact, translationally active ribonucleic acid. DNA, 2, 329–335.
Chang, S., Puryear, J., & Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter, 11(2), 113–116.
Chirgwin, J. M., Przybyla, A. E., MacDonald, R. J., & Rutter, W. J. (1979). Isolation of biologically active ribonucleic acid from sources enriched with ribonuclease. Biochemistry, 18, 5294–5299.
Chomczynski, P., & Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry, 162, 156–159.
Galau, G. A., Legocki, A. B., Greenway, S. C., & Dure, L. S. (1981). Cotton messenger RNA sequences exist in both polyadenylated and nonpolyadenylated forms. Journal of Biological Chemistry, 256, 2551–2560.
Geuna, F., Hartings, H., & Scienza, A. (1998). A new method for rapid extraction of high quality RNA from recalcitrant tissues of grapevine. Plant Molecular Biology Reporter, 16(1), 61–67.
Ghangal, R., Raghuvanshi, S., & Sharma, P. C. (2009). Isolation of good quality RNA from a medicinal plant seabuckthorn, rich in secondary metabolites. Plant Physiology and Biochemistry, 47, 1113–1115.
Han, J. H., Stratowa, C., & Rutter, W. J. (1987). Isolation of full-length putative rat lysophospholipase cDNA using improved methods for mRNA isolation and cDNA cloning. Biochemistry, 26, 1617–1625.
Khan, F., Islam, A., & Sathasivan, A. K. (2004). Rapid method for high quality RNA isolation from jute: Corchorus capsularis L. and Corchorus olitorius L. Plant Tissue Culture, 14(1), 63–68.
Lal, L., Sahoo, R., Gupta, R. K., Sharma, P., & Kumar, S. (2001). RNA isolation from high phenolic tea leaves and apical buds. Plant Molecular Biology Reporter, 19, 181–186.
Malnoy, M., Reynoird, J. P., Mourgues, F., Chevreau, E., & Simoneau, P. (2001). A method for isolating Total RNA from pear leaves. Plant Molecular Biology Reporter, 19(1), 69–74.
Mondal, R. (2000). Diversified jute products. Published by International Jute Organization (IJO) (pp. 115–118). Bangladesh: Dhaka.
Rouhibakhsh, A., Priya, J., Periasamy, M., Haq, Q. M. I., & Malathi, V. G. (2008). An improved DNA isolation method and PCR protocol for efficient detection of multicomponents of begomovirus in legumes. Journal of Virological Methods, 147, 37–42.
Salzman, R. A., Fujita, T., Zhu-Salzman, K., Hasegawa, P. M., & Bressan, R. A. (1999). An improved RNA isolation method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant Molecular Biology Reporter, 17, 11–17.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press.
Schultz, D. J., Craig, R., Cox-Foster, L. D., Mumma, O. R., & Medford, I. J. (2004). RNA isolation from recalcitrant plant tissue. Plant Molecular Biology Reporter, 12(4), 310–316.
Stephens, J., & Halpin, C. (2007). Lignin manipulation for fibre improvement. In: Improvement of crop plants for industrial end uses, P. Ranalli (Ed.), 129–153.
Taliaferro, J. M., Islam, A. S., & Sathasivan, K. (2006). Expressed sequence tags (ESTs) from a jute (Corchorus olitorius L.) cDNA library. Plant Tissue Culture and Biotechnology, 16(2), 95–104.
Valenzuela-Avendaño, J. P., Mota, E. I. A., Gabriel Lizama, U. C., Perera, S. R., Valenzuela-Soto, E. M., & Zúñiga Aguilar, J. J. (2005). Use of a simple method to isolate intact RNA from partially hydrated Selaginella Lepidophylla plants. Plant Molecular Biology Reporter, 23, 199–205.
Wan, C. Y., & Wilkins, T. A. (1994). A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Analytical Biochemistry, 223, 7–12.
Wang, X., Tian, W., & Li, Y. (2008). Development of an Efficient Protocol of RNA Isolation from Recalcitrant Tree Tissues. Molecular Biotechnology, 38, 57–64.
Acknowledgments
Financial assistance in the form the grant support to this laboratory from the Department of Biotechnology, Government of India is acknowledged. The authors would like to thank Dr. P. Paria for valuable suggestions in the manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Pradipta Samanta, Sanjoy Sadhukhan, both the authors have equal contribution.
An erratum to this article can be found at http://dx.doi.org/10.1007/s12033-011-9424-4
Rights and permissions
About this article
Cite this article
Samanta, P., Sadhukhan, S., Das, S. et al. Isolation of RNA from Field-Grown Jute (Corchorus capsularis) Plant in Different Developmental Stages for Effective Downstream Molecular Analysis. Mol Biotechnol 49, 109–115 (2011). https://doi.org/10.1007/s12033-011-9376-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9376-8