Abstract
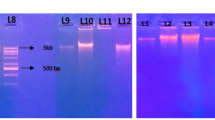
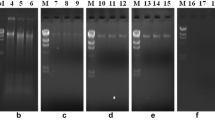
A new method is described for extraction of metagenomic DNA from soil and sediments which is based on DNA adsorption to silica without the use of phenol, ethanol precipitation or a cesium chloride gradient. High-quality DNA was obtained, and PCR inhibition was overcome by adding bovine serum albumin and adjusting magnesium concentration. By using PCR-DGGE with Firmicutes and lactic acid bacteria-specific primers the extracted metagenomic DNA was shown to contain a mixture of bacterial genomes. This method can be used for screening bacterial diversity in soil and sediment samples.



Similar content being viewed by others
References
Bissett, A., Bowman, J., & Burke, C. (2006). Bacterial diversity in organically enriched fish farm sediments. FEMS Microbiology Ecology, 55, 48–56.
Head, I., Saunders, J., & Pickup, R. (1998). Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microbial Ecology, 35, 1–21.
Muyzer, G., de Waal, E. C., & Uitterlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology, 59, 695–700.
Tebbe, C., & Vahjen, W. (1993). Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Applied and Environmental Microbiology, 59, 2657–2665.
Leff, L., Dana, J., McArthur, J., & Shimkets, L. (1995). Comparison of methods of DNA extraction from stream sediments. Applied and Environmental Microbiology, 61, 1141–1143.
Tsai, Y., & Olson, B. (1992). Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Applied and Environmental Microbiology, 58, 2292–2295.
Roose-Amsaleg, C., Garnier-Sillam, E., & Harry, M. (2001) Extraction and purification of microbial DNA from soil and sediments samples. Applied Soil Ecology, 18, 47–60.
Jacobsen, C. S., & Rasmussen, O. F. (1992). Development and application of a new method to extract bacterial DNA from soil based on separation of bacteria from soil with cation-exchange resin. Applied and Environmental Microbiology, 58, 2458–2462.
Picard, C., Ponsonnet, C., Paget, E., Nesme, X., & Simonet, P. (1992). Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Applied and Environmental Microbiology, 58, 2717–2722.
Porteous, L. A., & John, L. A. (1993). A simple mini-method to extract DNA directly from soil for use with polymerase chain reaction amplification. Current Microbiology 27, 115–118.
Arbeli, Z., & Fuentes, C. L. (2007). Improved purification and PCR amplification of DNA from environmental samples. FEMS Microbiology Letters, 272, 269–275.
Miller, D., Bryant, J., Madsen, E., & Ghiorse, W. (1999). Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Applied and Environmental Microbiology, 65, 4715–4724.
Zhou, J., Bruns, M. A., & Tiedje, J. M. (1996). DNA recovery from soils of diverse composition. Applied and Environmental Microbiology, 62, 316–322.
Gray, J., & Herwig, R. (1996). Phylogenetic analysis of the bacterial communities in marine sediments. Applied and Environmental Microbiology, 62, 4049–4059.
Kreader, C. A. (1996). Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Applied and Environmental Microbiology, 62, 1102–1106.
Sharma, P., Capalash, N., & Kaur, J. (2007). An improved method for single step purification of metagenomic DNA. Molecular Biotechnology, 36, 61–63.
Pedersen, C., Jonsson, H., Linberg, J., & Roos, S. (2004). Microbiological characterization of wet wheat distillers’ grain, with focus on isolation of lactobacilli with potencial as probotic. Applied and Environmental Microbiology, 70, 1522–1527.
Tsai, Y. L., & Olson, B. H. (1991). Rapid method for direct extraction of DNA from soil and sediments. Applied and Environmental Microbiology, 57, 1070–1074.
Acknowledgments
This research was supported by the “Fondo Sectorial de Investigación Ambiental” from Mexico, Project FOSEMARNAT-2004-01-280.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rojas-Herrera, R., Narváez-Zapata, J., Zamudio-Maya, M. et al. A Simple Silica-based Method for Metagenomic DNA Extraction from Soil and Sediments. Mol Biotechnol 40, 13–17 (2008). https://doi.org/10.1007/s12033-008-9061-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-008-9061-8