Abstract
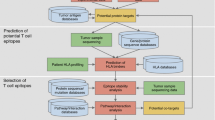
Pancreatic cancer is a highly aggressive and often lethal malignancy with limited treatment options. Its late-stage diagnosis and resistance to conventional therapies make it a significant challenge in oncology. Immunotherapy, particularly cancer vaccines, has emerged as a promising avenue for treating pancreatic cancer. Multi-epitope vaccines, designed to target multiple epitopes derived from various antigens associated with pancreatic cancer, have gained attention as potential candidates for improving therapeutic outcomes. In this study, we have explored transcriptomics and protein expression databases to identify potential upregulated proteins in pancreatic cancer cells. After examining a total of 21,054 proteins from various databases, it was discovered that 143 proteins expressed differently in malignant and healthy cells. The CTL, HTL and BCE epitopes were predicted for the shortlisted proteins. 51,840 vaccine constructs were created by concatenating CTL, HTL, and B-cell epitopes in the respective sequences. The best 86 structures were selected from a set of 51,840 designs after they were analyzed for vaxijenicity, allergenicity, toxicity, and antigenicity scores. In further simulation of the immune response using constructs, it was found that 41417, 37961, and 40841 constructs could produce a strong immune response when injected. Further, it was found that construct 37961 showed stronger interaction and stability with TLR-9 as determined from the large-scale molecular dynamics simulations. Moreover, the 37961 construct has shown interactions with TLR-9 suggests its potential in inducing immune response. In addition, construct 37961 has shown 100% predicted solubility in the E. coli expression system. Overall, the study indicates the designed construct 37961 has the potential to induce an anti-tumor immune response and long-standing protection pending further studies.
Graphical abstract





Similar content being viewed by others
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48.
Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–6.
Hayashi A, Hong J, Iacobuzio-Donahue CA. The pancreatic cancer genome revisited. Nat Rev Gastroenterol Hepatol. 2021;18(7):469–81.
Gheorghe G, Diaconu CC, Ionescu V, Constantinescu G, Bacalbasa N, Bungau S, et al. Risk factors for pancreatic cancer: emerging role of viral hepatitis. J Personal Med. 2022;12(1):83.
Wang DS, Chen DL, Ren C, Wang ZQ, Qiu MZ, Luo HY, et al. ABO blood group, hepatitis B viral infection and risk of pancreatic cancer. Int J Cancer. 2012;131(2):461–8.
Saung MT, Zheng L. Current standards of chemotherapy for pancreatic cancer. Clin Ther. 2017;39(11):2125–34.
Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–25.
Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–703.
Pontén F, Jirström K, Uhlen M. The human protein atlas—a tool for pathology. J Pathol: J Pathol Soc Great Br Irel. 2008;216(4):387–93.
Papatheodorou I, Fonseca NA, Keays M, Tang YA, Barrera E, Bazant W, et al. Expression atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018;46(D1):D246–51.
Zeng J, Zhang Y, Shang Y, Mai J, Shi S, Lu M, et al. CancerSCEM: a database of single-cell expression map across various human cancers. Nucleic Acids Res. 2022;50(D1):D1147–55.
Tang G, Cho M, Wang X. OncoDB: an interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022;50(D1):D1334–9.
Thumuluri V, Almagro Armenteros JJ, Johansen AR, Nielsen H, Winther O. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022;50(W1):W228–34.
Hallgren J, Tsirigos KD, Pedersen MD, Almagro Armenteros JJ, Marcatili P, Nielsen H, et al. (2022) DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv. 2022:2022.04. 08.487609
Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8(1):1–9.
Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48(W1):W449–54.
Clifford JN, Høie MH, Deleuran S, Peters B, Nielsen M, Marcatili P. BepiPred-3.0: improved B-cell epitope prediction using protein language models. Protein Sci. 2022;31(12):e4497.
Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276(1–2):172–4.
Dimitrov I, Flower DR, Doytchinova I. AllerTOP-a server for in silico prediction of allergens. BMC Bioinform. 2013. https://doi.org/10.1186/1471-2105-14-S6-S4.
Morozov V, Rodrigues CH, Ascher DB. CSM-toxin: a web-server for predicting protein toxicity. Pharmaceutics. 2023;15(2):431.
Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010;5(4):e9862.
Stolfi P, Castiglione F, Mastrostefano E, Di Biase I, Di Biase S, Palmieri G, et al. In-silico evaluation of adenoviral COVID-19 vaccination protocols: assessment of immunological memory up to 6 months after the third dose. Front Immunol. 2022;13: 998262.
Ragone C, Manolio C, Cavalluzzo B, Mauriello A, Tornesello ML, Buonaguro FM, et al. Identification and validation of viral antigens sharing sequence and structural homology with tumor-associated antigens (TAAs). J Immunother Cancer. 2021. https://doi.org/10.1136/jitc-2021-002694.
Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19(6):679–82.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–9.
Vaz J, Andersson R. Intervention on toll-like receptors in pancreatic cancer. World J Gastroenterol: WJG. 2014;20(19):5808.
Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12(2):255–78.
Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–23.
Krüger DM, Ahmed A, Gohlke H. NMSim web server: integrated approach for normal mode-based geometric simulations of biologically relevant conformational transitions in proteins. Nucleic Acids Res. 2012;40(W1):W310–6.
Agostini F, Cirillo D, Livi CM, Delli Ponti R, Tartaglia GG. cc SOL omics: a webserver for solubility prediction of endogenous and heterologous expression in Escherichia coli. Bioinformatics. 2014;30(20):2975–7.
Grossberg AJ, Chu LC, Deig CR, Fishman EK, Hwang WL, Maitra A, et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin. 2020;70(5):375–403.
Sigalov AB. New therapeutic strategies targeting transmembrane signal transduction in the immune system. Cell Adh Migr. 2010;4(2):255–67.
Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–10.
Tyka MD, Keedy DA, André I, DiMaio F, Song Y, Richardson DC, et al. Alternate states of proteins revealed by detailed energy landscape mapping. J Mol Biol. 2011;405(2):607–18.
Gasteiger E, Hoogland C, Gattiker A, Se D, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. Totowa: Springer; 2005.
Kara EE, Comerford I, Fenix KA, Bastow CR, Gregor CE, McKenzie DR, et al. Tailored immune responses: novel effector helper T cell subsets in protective immunity. PLoS Pathog. 2014;10(2): e1003905.
Finn OJ. Cancer vaccines: between the idea and the reality. Nat Rev Immunol. 2003;3(8):630–41.
Luo W, Yin Q. B cell response to vaccination. Immunol Invest. 2021;50(7):780–801.
Zahavi D, Weiner L. Monoclonal antibodies in cancer therapy. Antibodies. 2020;9(3):34.
Nardin A, Abastado J-P. Macrophages and cancer. Front Biosci. 2008;13(3):494–505.
Burke JD, Young HA. IFN-γ: a cytokine at the right time, is in the right place. Semin Immunol. 2019. https://doi.org/10.1016/j.smim.2019.05.002.
Helminen O, Huhta H, Kauppila JH, Lehenkari PP, Saarnio J, Karttunen TJ. Localization of nucleic acid-sensing toll-like receptors in human and mouse pancreas. APMIS. 2017;125(2):85–92.
Pahlavanneshan S, Sayadmanesh A, Ebrahimiyan H, Basiri M. Toll-like receptor-based strategies for cancer immunotherapy. J Immunol Res. 2021;2021:1–14.
Ruan M, Thorn K, Liu S, Li S, Yang W, Zhang C, et al. The secretion of IL-6 by CpG-ODN-treated cancer cells promotes T-cell immune responses partly through the TLR-9/AP-1 pathway in oral squamous cell carcinoma. Int J Oncol. 2014;44(6):2103–10.
Connor AA, Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat Rev Cancer. 2022;22(3):131–42.
Pilla L, Rivoltini L, Patuzzo R, Marrari A, Valdagni R, Parmiani G. Multipeptide vaccination in cancer patients. Expert Opin Biol Ther. 2009;9(8):1043–55.
Ray SK, Mukherjee S. Altering landscape of cancer vaccines: unique platforms, research on therapeutic applications and recent patents. Recent Pat Anti-Cancer Drug Discovery. 2023;18(2):133–46.
Gan L-L, Hii L-W, Wong S-F, Leong C-O, Mai C-W. Molecular mechanisms and potential therapeutic reversal of pancreatic cancer-induced immune evasion. Cancers. 2020;12(7):1872.
de Paula PL, da Luz FAC, dos Anjos PB, Brigido PC, de Araujo RA, Goulart LR, et al. Peptide vaccines in breast cancer: the immunological basis for clinical response. Biotechnol Adv. 2015;33(8):1868–77.
Tamiola K, Acar B, Mulder FA. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc. 2010;132(51):18000–3.
Saber MM, Monir N, Awad AS, Elsherbiny ME, Zaki HF. TLR9: a friend or a foe. Life Sci. 2022;307:120874.
Lin X, Ye L, Wang X, Liao Z, Dong J, Yang Y, et al. Follicular helper T cells remodel the immune microenvironment of pancreatic cancer via secreting CXCL13 and IL-21. Cancers. 2021;13(15):3678.
den Haan JM, Arens R, van Zelm MC. The activation of the adaptive immune system: cross-talk between antigen-presenting cells, T cells and B cells. Immunol Lett. 2014;162(2):103–12.
Alshaker HA, Matalka KZ. IFN-γ, IL-17 and TGF-β involvement in shaping the tumor microenvironment: the significance of modulating such cytokines in treating malignant solid tumors. Cancer Cell Int. 2011;11(1):1–12.
Funding
SB acknowledges the National Institute of Technology Warangal for funding in the form of postdoctoral fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare there is no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file3 (MP4 44487 KB)
12032_2024_2334_MOESM5_ESM.jpg
Figure S1. Top vaccine constructs modelled structures with their corresponding Ramachandran plots. (A) 41417, (B) 40841, (C) 37385, (D) 37962, (E) 37937, (F) 37997, (G) 37386, (H) 51785 and (I) 37097. All of the constructs contains large random coil content and most residues are in disallowed regions. Supplementary file5 (JPG 6262 KB)
12032_2024_2334_MOESM6_ESM.jpg
Figure S2. Vaccine constructs solubility assessment for (A) 41417, (B) 40841, and (C) 37385 constructs determined from CCSol server. All of the constructs are highly soluble in E. coli system. Supplementary file6 (JPG 1453 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Banesh, S., Patil, N., Chethireddy, V.R. et al. Design and evaluation of a multiepitope vaccine for pancreatic cancer using immune-dominant epitopes derived from the signature proteome in expression datasets. Med Oncol 41, 90 (2024). https://doi.org/10.1007/s12032-024-02334-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-024-02334-4