Abstract
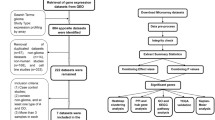
The abnormal expressions of enhancer-associated genes have been reported to be correlated with poor prognosis of tumors, including glioblastoma (GBM). The objective of the current study is to predict prognosis by identifying enhancer-associated genes (EAGs). The profiles of genome-wide expressions of low-grade glioma (LGG) and GBM tissues in The Cancer Genome Atlas (TCGA) dataset were obtained to explore the expression patterns of EAGs in diffuse glioma. The capacity of prognosis prediction was validated by Rembrandt and GSE16011. Moreover, qPCR was utilized to confirm the effect of JQ1 and THZ1 on the EAGs. We detected 35 differentially expressed EAGs, which were predictive of overall survival. These candidate EAGs were then subjected to the multivariate cox regression analysis and were further scoped down to four signature genes, including TRAM2, SMAGP, KDELC2, and C7ORF25. A total of 662 patients were then stratified according to the expression levels of these four signature genes. The high-risk group accounted for poorer prognosis based on the Rembrandt and GSE16011 databases. The results of qPCR also demonstrated that the expression of the four EAGs could be abolished by JQ1 (bromodomain inhibitor) and THZ1 (CDK7 inhibitor) treatment. Our study not only highlights the potential role of EAGs, which can be used to improve clinical prognosis prediction in patients with diffuse glioma, but also sheds light on the specific biomarkers and therapeutic targets for diffuse glioma patients.







Similar content being viewed by others
References
Bahr C et al (2018) A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nat 553(7689):515–520
Cai L, Kirchleitner SV, Zhao D, Li M, Tonn JC, Glass R, Kalin RE (2020) Glioblastoma exhibits inter-individual heterogeneity of TSPO and LAT1 expression in neoplastic and parenchymal cells. Int J Mol Sci 21(2):612. https://doi.org/10.3390/ijms21020612
Carlsson SK, Brothers SP, Wahlestedt C (2015) Emerging treatment strategies for glioblastoma multiforme. Embo Mol Med 6(11):1359–1370
Cole PA (2008) Chemical probes for histone-modifying enzymes. Nat Chem Biol 4(10):590–597
Dave K et al (2017) Mice deficient ofMycsuper-enhancer region reveal differential control mechanism between normal and pathological growth. Elife 6:e23382
Dawson MA et al (2011) Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nat 478(7370):529–533
Delmore J et al (2011) BET bromodomain inhibition asa therapeutic strategy to target c-Myc. Cell 146(6):904–917
Dolecek TA, Propp JM, Stroup NE, Kruchko C (2013) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States. In: 2005–2009 Neuro-oncology, 14 Suppl 5:v1
Duan Q et al (2016) Super enhancers at the miR-146a and miR-155 genes contribute to self-regulation of inflammation. Biochem Biophys Acta 4:564–571
Elsässer SJ, Allis CD, Lewis PW (2011) New Epigenetic Drivers of Cancers Sci 331(6021):1145–1146
Feinberg AP, Tycko B (2004) The history of cancer epigenetics. Nat Rev Cancer 4(2):143–153
Fukuoka K et al (2019) BRAF V600E mutant oligodendroglioma-like tumors with chromosomal instability in adolescents and young adults. Brain Pathol 30(3):515–523. https://doi.org/10.1111/bpa.12799
Furnari FB et al (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21(21):2683–2710
Gelato K et al (2017) Super-enhancers define a proliferative PGC-1α-expressing melanoma subgroup sensitive to BET inhibition. Oncog 37(4):512–521
Geutjes EJ, Bajpe PK, Bernards R (2012) Targeting the epigenome for treatment of cancer. Oncog 31(34):3827–3844
Giniger E, Ptashne M (1988) Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci USA 85(2):382–386
Griggs DW, Johnston M (1991) Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci U S A 88(19):8597–8601
Hnisz D et al (2013) Super-enhancers in the control of cell identity and disease. Cell 155(4):934–947
Issa JP, Kantarjian HM (2009) Targeting DNA methylation Clinical Cancer Research. Off J Am Assoc Cancer Res 15:3938
Jiang YY et al (2017) Targeting super-enhancer-associated oncogenes in oesophageal squamous cell carcinoma. Gut 66(8):1358–1368
Johnson KC, Houseman EA, King JE, von Herrmann KM, Fadul CE, Christensen BC (2016) 5-Hydroxymethylcytosine localizes to enhancer elements and is associated with survival in glioblastoma patients. Nat Commun 7:13177. https://doi.org/10.1038/ncomms13177
Li P, Wu M (2017) Epigenetic mechanisms of glioblastoma. Exon Publications 43–58
Lieberman F (2017) Glioblastoma update: molecular biology, diagnosis, treatment, response assessment, and translational clinical trials. F1000research 6:1892
Louis DN et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Lovén J et al (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153(2):320–334
Ludwig K, Kornblum HI (2017) Molecular Markers in Glioma J Neuro-Oncol 134:1–8
Mahase S, Rattenni RN, Wesseling P, Leenders W, Baldotto C, Jain R, Zagzag D (2017) Hypoxia-mediated mechanisms associated with antiangiogenic treatment resistance in glioblastomas. Am J Pathol 187:940
Marks PA, Xu WS (2010) Histone deacetylase inhibitors: potential in cancer therapy. J Cell Biochem 107(4):600–608
Meng W et al (2018a) Enhanced efficacy of histone deacetylase inhibitor combined with bromodomain inhibitor in glioblastoma. J Exp Clin Cancer Res 37:241. https://doi.org/10.1186/s13046-018-0916-y
Meng W et al (2018b) CDK7 inhibition is a novel therapeutic strategy against GBM both in vitro and in vivo. Cancer Manag Res 10:5747–5758. https://doi.org/10.2147/CMAR.S183696
Nagaraja S et al (2017) Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 31(5):635–652
Ostrom QT et al (2014) The epidemiology of glioma in adults: a “state of the science” review. Neuro-Oncol 16(7):896–913
Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS (2019) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol 21:v1–v100. https://doi.org/10.1093/neuonc/noz150
Pott S, Lieb JD (2015) What are super-enhancers? Nat Genet 47(1):8–12
Sathornsumetee S, Reardon DA, Desjardins A, Quinn JA, Vredenburgh JJ, Rich JN (2010) Molecularly targeted therapy for malignant glioma. Cancer 110(1):13–24
Sengupta S, George RE (2017) Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer 3(4):269–281
Sim HW, Morgan ER, Mason WP (2017) Contemporary management of high-grade gliomas. Cns Oncol 7(01):51–65
Stoyanov GS, Dzhenkov D, Ghenev P, Iliev B, Enchev Y, Tonchev AB (2018) Cell biology of glioblastoma multiforme: from basic science to diagnosis and treatment. Med Oncol 35:27
Umehara T et al (2019) Distribution differences in prognostic copy number alteration profiles in IDH-wild-type glioblastoma cause survival discrepancies across cohorts. Acta Neuropathol Commun 7:99. https://doi.org/10.1186/s40478-019-0749-8
Walid MS (2008) Prognostic factors for long-term survival after glioblastoma. Permanente Journal 12:45
Wang L, Hu G (2016) Remodeling super-enhancers and oncogenic transcription. Cell Cycle 15(23):3157–3158
Weinstein CLJ et al (2020) A phase 3 study evaluating the safety and efficacy of a pediatric dose of mometasone furoate with and without formoterol for persistent asthma. Pediatr Pulmonol 55(4):882–889. https://doi.org/10.1002/ppul.24667
Whyte W et al (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153(2):307–319
You JS, Jones PA (2012) Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22(1):9–20
Yuan C et al (2017) Super enhancer associated RAI14 is a new potential biomarker in lung adenocarcinoma. Oncotarget 8(62):105251–105261
Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J (2017) An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol Neurobiol 55(5):3684–3697
Funding
This work was funded by National Natural Science Foundation of China (no. 81271382).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
†Jiajia Wang and Xin Ma contributed equally to this work
Rights and permissions
About this article
Cite this article
Wang, J., Ma, X. & Ma, J. Identification of Four Enhancer-Associated Genes as Risk Signature for Diffuse Glioma Patients. J Mol Neurosci 72, 410–419 (2022). https://doi.org/10.1007/s12031-021-01861-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-021-01861-3