Abstract
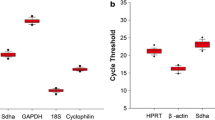
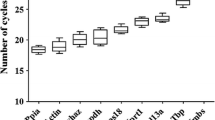
Sudden infant death syndrome (SIDS) is the main cause of post-neonatal infant death in most developed countries. It is still of ambiguous etiology. Gene expression studies of relevant target genes using reverse transcription quantitative real-time PCR (RT-qPCR) in SIDS cases, and comparing them with age-matched controls, could help in understanding the pathogenesis of SIDS. However, selecting inadequate reference genes used for normalization of the RT-qPCR gene expression data can give misleading results. The aim of the present study was to identify reference genes with the most stable expression in post-mortem brainstem samples of SIDS and control cases. Among the five candidate reference genes (GAPDH, GUSB, HMBS, SDHA, UBXN6) studied in both groups, SDHA and UBXN6 were identified as the most stable. To further demonstrate the importance of using validated genes for RT-qPCR data normalization, the expression of a potential gene of interest in SIDS, the RPS27A gene, was evaluated using validated versus non-validated reference genes for normalization. This gene encodes the ubiquitin protein that has been shown in other pathological studies to be induced in SIDS. Using the identified most stable genes for normalization of RPS27A gene expression data revealed, as expected, a statistically significant up-regulation in SIDS as compared to the controls. However, using a single unstable reference gene for normalization resulted in no significant differences in transcript abundance of RPS27A between SIDS and the controls. This emphasizes the need for validation of the suitability of reference genes used in a given tissue type under certain experimental conditions.




Similar content being viewed by others
References
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034.
Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6(4):279–84.
Koppelkamm A, Vennemann B, Fracasso T, Lutz-Bonengel S, Schmidt U, Heinrich M. Validation of adequate endogenous reference genes for the normalisation of qPCR gene expression data in human post mortem tissue. Int J Legal Med. 2010;124(5):371–80.
Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:9.
Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54(4):391–406.
Bemeur C, Ste-Marie L, Desjardins P, Hazell AS, Vachon L, Butterworth R, et al. Decreased beta-actin mRNA expression in hyperglycemic focal cerebral ischemia in the rat. Neurosci Lett. 2004;357(3):211–4.
Bas A, Forsberg G, Hammarstrom S, Hammarstrom ML. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol. 2004;59(6):566–73.
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37(1):112–4, 6, 8–9.
Huth A, Vennemann B, Fracasso T, Lutz-Bonengel S, Vennemann M. Apparent versus true gene expression changes of three hypoxia-related genes in autopsy derived tissue and the importance of normalisation. Int J Legal Med. 2013;127(2):335–44.
Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29(2):332–7.
Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, et al. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75(2–3):291–5.
Heinrich M, Lutz-Bonengel S, Matt K, Schmidt U. Real-time PCR detection of five different “endogenous control gene” transcripts in forensic autopsy material. Forensic Sci Int Genet. 2007;1(2):163–9.
Preece P, Virley DJ, Costandi M, Coombes R, Moss SJ, Mudge AW, et al. An optimistic view for quantifying mRNA in post-mortem human brain. Brain Res Mol Brain Res. 2003;116(1–2):7–16.
Krous HF, Beckwith JB, Byard RW, Rognum TO, Bajanowski T, Corey T, et al. Sudden infant death syndrome and unclassified sudden infant deaths: a definitional and diagnostic approach. Pediatrics. 2004;114(1):234–8.
Kinney HC, Thach BT. The sudden infant death syndrome. N Engl J Med. 2009;361(8):795–805.
Carpenter RG, Irgens LM, Blair PS, England PD, Fleming P, Huber J, et al. Sudden unexplained infant death in 20 regions in Europe: case control study. Lancet. 2004;363(9404):185–91.
Vennemann MM, Findeisen M, Butterfass-Bahloul T, Jorch G, Brinkmann B, Kopcke W, et al. Modifiable risk factors for SIDS in Germany: results of GeSID. Acta Paediatr. 2005;94(6):655–60.
Hunt CE, Darnall RA, McEntire BL, Hyma BA. Assigning cause for sudden unexpected infant death. Forensic Sci Med Pathol. 2015. doi:10.1007/s12024-014-9650-8.
Courts C, Madea B. Genetics of the sudden infant death syndrome. Forensic Sci Int. 2010;203(1–3):25–33.
Klintschar M, Heimbold C. Association between a functional polymorphism in the MAOA gene and sudden infant death syndrome. Pediatrics. 2012;129(3):e756–61.
Millat G, Kugener B, Chevalier P, Chahine M, Huang H, Malicier D, et al. Contribution of long-QT syndrome genetic variants in sudden infant death syndrome. Pediatr Cardiol. 2009;30(4):502–9.
Paterson DS, Trachtenberg FL, Thompson EG, Belliveau RA, Beggs AH, Darnall R, et al. Multiple serotonergic brainstem abnormalities in sudden infant death syndrome. JAMA. 2006;296(17):2124–32.
Opdal SH, Rognum TO. Gene variants predisposing to SIDS: current knowledge. Forensic Sci Med Pathol. 2011;7(1):26–36.
Casale V, Oneda R, Matturri L, Lavezzi AM. Investigation of 5-HTT expression using quantitative real-time PCR in the human brain in SIDS Italian cases. Exp Mol Pathol. 2013;94(1):239–42.
Quan L, Zhu BL, Ishida K, Oritani S, Taniguchi M, Fujita MQ, et al. Intranuclear ubiquitin immunoreactivity of the pigmented neurons of the substantia nigra in fatal acute mechanical asphyxiation and drowning. Int J Legal Med. 2001;115(1):6–11.
Toupalik P, Bouska I. Immunohistochemical findings in the central nervous system in sudden infant death. Soud Lek. 1999;44(2):17–20.
Sawaguchi T, Patricia F, Kadhim H, Groswasser J, Sottiaux M, Nishida H, et al. The correlation between ubiquitin in the brainstem and sleep apnea in SIDS victims. Early Hum Dev. 2003;75(Suppl):S75–86.
Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int J Legal Med. 2012;126(6):943–52.
Barrachina M, Castano E, Ferrer I. TaqMan PCR assay in the control of RNA normalization in human post-mortem brain tissue. Neurochem Int. 2006;49(3):276–84.
Coulson DT, Brockbank S, Quinn JG, Murphy S, Ravid R, Irvine GB, et al. Identification of valid reference genes for the normalization of RT qPCR gene expression data in human brain tissue. BMC Mol Biol. 2008;9:46.
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22.
Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50(4):227–30.
Demendi C, Borzsonyi B, Nagy ZB, Rigo J Jr, Pajor A, Joo JG. Gene expression patterns of insulin-like growth factor 1, 2 (IGF-1, IGF-2) and insulin-like growth factor binding protein 3 (IGFBP-3) in human placenta from preterm deliveries: influence of additional factors. Eur J Obstet Gynecol Reprod Biol. 2012;160(1):40–4.
Neary MT, Breckenridge RA. Hypoxia at the heart of sudden infant death syndrome? Pediatr Res. 2013;74(4):375–9.
Rognum TO, Saugstad OD. Hypoxanthine levels in vitreous humor: evidence of hypoxia in most infants who died of sudden infant death syndrome. Pediatrics. 1991;87(3):306–10.
Jones KL, Krous HF, Nadeau J, Blackbourne B, Zielke HR, Gozal D. Vascular endothelial growth factor in the cerebrospinal fluid of infants who died of sudden infant death syndrome: evidence for antecedent hypoxia. Pediatrics. 2003;111(2):358–63.
Schoor O, Weinschenk T, Hennenlotter J, Corvin S, Stenzl A, Rammensee HG, et al. Moderate degradation does not preclude microarray analysis of small amounts of RNA. Biotechniques. 2003;35(6):1192–6, 8–201.
Stan AD, Ghose S, Gao XM, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ, et al. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123(1):1–11.
Durrenberger PF, Fernando S, Kashefi SN, Ferrer I, Hauw JJ, Seilhean D, et al. Effects of antemortem and postmortem variables on human brain mRNA quality: a BrainNet Europe study. J Neuropathol Exp Neurol. 2010;69(1):70–81.
Gomez-Nicola D, Boche D. Post-mortem analysis of neuroinflammatory changes in human Alzheimer’s disease. Alzheimers Res Ther. 2015;7(1):42.
Webster MJ. Tissue preparation and banking. Prog Brain Res. 2006;158:3–14.
Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Asp Med. 2006;27(2–3):126–39.
Antonov J, Goldstein DR, Oberli A, Baltzer A, Pirotta M, Fleischmann A, et al. Reliable gene expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab Invest. 2005;85(8):1040–50.
van der Linden A, Blokker BM, Kap M, Weustink AC, Riegman PH, Oosterhuis JW. Post-mortem tissue biopsies obtained at minimally invasive autopsy: an RNA-quality analysis. PLoS ONE. 2014;9(12):e115675.
Birdsill AC, Walker DG, Lue L, Sue LI, Beach TG. Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 2011;12(4):311–8.
Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55(4):346–52.
Preece P, Cairns NJ. Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Brain Res Mol Brain Res. 2003;118(1–2):60–71.
Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57(5):549–58.
Moon RY. SIDS and other sleep-related infant deaths: expansion of recommendations for a safe infant sleeping environment. Pediatrics. 2011;128(5):1030–9.
Acknowledgments
We gratefully thank Ruth Volland for assistance in the SPSS software.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
El-Kashef, N., Gomes, I., Mercer-Chalmers-Bender, K. et al. Validation of adequate endogenous reference genes for reverse transcription-qPCR studies in human post-mortem brain tissue of SIDS cases. Forensic Sci Med Pathol 11, 517–529 (2015). https://doi.org/10.1007/s12024-015-9717-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12024-015-9717-1