Abstract
Background
Heterotopic ossification (HO) is one of the most intractable conditions following injury to the musculoskeletal system. In recent years, much attention has been paid to the role of lncRNA in musculoskeletal disorders, but its role in HO was still unclear. Therefore, this study attempted to determine the role of lncRNA MEG3 in the formation of post-traumatic HO and further explore the underlying mechanisms.
Results
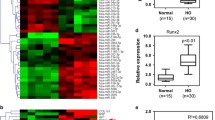
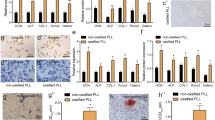
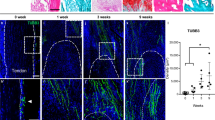
On the basis of high-throughput sequencing and qPCR validation, elevated expression of the lncRNA MEG3 was shown during traumatic HO formation. Accordingly, in vitro experiments demonstrated that lncRNA MEG3 promoted aberrant osteogenic differentiation of tendon-derived stem cells (TDSCs). Mechanical exploration through RNA pulldown, luciferase reporter gene assay and RNA immunoprecipitation assay identified the direct binding relationship between miR-129-5p and MEG3, or miR-129-5p and TCF4. Further rescue experiments confirmed the miR-129-5p/TCF4/β-catenin axis to be downstream molecular cascade responsible for the osteogenic-motivating effects of MEG3 on the TDSCs. Finally, experiments in a mouse burn/tenotomy model corroborated the promoting effects of MEG3 on the formation of HO through the miR-129-5p/TCF4/β-catenin axis.
Conclusions
Our study demonstrated that the lncRNA MEG3 promoted osteogenic differentiation of TDSCs and thus the formation of heterotopic ossification, which could be a potential therapeutic target.








Similar content being viewed by others
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- HO:
-
Heterotopic ossification
- TDSCs:
-
Tendon stem cells
- BMSCs:
-
Bone marrow mesenchymal cells
- MEG3:
-
Maternally expressed gene 3
- TCF4:
-
Transcription factor 4
- FBS:
-
Fetal Bovine Serum
- PDGFRα:
-
Platelet-derived growth factor receptor-α
- EDTA:
-
Ethylenediaminetetraacetic acid
- FISH :
-
Fluorescence in situ hybridization
- IHC:
-
Immunohistochemistry
- micro-CT:
-
Micro-computed tomography
- ECL:
-
Electrochemiluminescence
- BCA:
-
Bicinchoninic acid
- TBST:
-
Tris buffered saline tween
- AAV9:
-
Adeno-associated virus 9
- ALP:
-
Alkaline phosphatase
- ARS:
-
Alizarin Red S
- RIP:
-
RNA Immunoprecipitation
- HE:
-
Hematoxylin and eosin
- PBS:
-
Phosphate-buffered saline
- qRT-PCR:
-
Quantitative reserve-transcriptase polymerase chain reaction
- SD:
-
Standard deviation
References
Feng, H., Xing, W., Han, Y., Sun, J., Kong, M., Gao, B., et al. (2020). Tendon-derived cathepsin K-expressing progenitor cells activate Hedgehog signaling to drive heterotopic ossification. The Journal of Clinical Investigation, 130(12), 6354–6365.
Agarwal, S., Loder, S. J., Breuler, C., Li, J., Cholok, D., Brownley, C., et al. (2017). Strategic Targeting of Multiple BMP Receptors Prevents Trauma-Induced Heterotopic Ossification. Molecular Therapy, 25(8), 1974–1987.
Schneider, J. C., Simko, L. C., Goldstein, R., Shie, V. L., Chernack, B., Levi, B., et al. (2017). Predicting Heterotopic Ossification Early After Burn Injuries: A Risk Scoring System. Annals of Surgery, 266(1), 179–184.
Wong, K. R., Mychasiuk, R., O’Brien, T. J., Shultz, S. R., McDonald, S. J., & Brady, R. D. (2020). Neurological heterotopic ossification: Novel mechanisms, prognostic biomarkers and prophylactic therapies. Bone Res., 8(1), 42.
Rudiger, H. A., Dittrich, M., Robinson, J., Mansour, T., Schwab, T., Stadelmann, V. A., et al. (2020). The Impact of Heterotopic Ossification on Self-Reported Outcomes After Total Hip Arthroplasty Using the Direct Anterior Approach. Journal of Bone and Joint Surgery. American Volume, 102(Suppl 2), 91–98.
Barfield, W. R., Holmes, R. E., & Hartsock, L. A. (2017). Heterotopic Ossification in Trauma. Orthopedic Clinics of North America, 48(1), 35–46.
Legosz, P., Drela, K., Pulik, L., Sarzynska, S., & Maldyk, P. (2018). Challenges of heterotopic ossification-Molecular background and current treatment strategies. Clinical and Experimental Pharmacology and Physiology, 45(12), 1229–1235.
Kodde, I. F., van Rijn, J., van den Bekerom, M. P., & Eygendaal, D. (2013). Surgical treatment of post-traumatic elbow stiffness: A systematic review. Journal of Shoulder and Elbow Surgery, 22(4), 574–580.
Lee, E. K., Namdari, S., Hosalkar, H. S., Keenan, M. A., & Baldwin, K. D. (2013). Clinical results of the excision of heterotopic bone around the elbow: A systematic review. Journal of Shoulder and Elbow Surgery, 22(5), 716–722.
Ramm Sander, P., Hau, P., Koch, S., Schutze, K., Bogdahn, U., Kalbitzer, H. R., et al. (2013). Stem cell metabolic and spectroscopic profiling. Trends in Biotechnology, 31(3), 204–213.
Pulik, L., Mierzejewski, B., Ciemerych, M. A., Brzoska, E., & Legosz, P. (2020). The survey of cells responsible for heterotopic ossification development in skeletal muscles-human and mouse models. Cells, 9(6), 1324.
Agarwal, S., Loder, S., Cholok, D., Li, J., Breuler, C., Drake, J., et al. (2017). Surgical Excision of Heterotopic Ossification Leads to Re-Emergence of Mesenchymal Stem Cell Populations Responsible for Recurrence. Stem Cells Translational Medicine, 6(3), 799–806.
Batista, P. J., & Chang, H. Y. (2013). Long noncoding RNAs: Cellular address codes in development and disease. Cell, 152(6), 1298–1307.
Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nature Medicine, 21(11), 1253–1261.
Fatica, A., & Bozzoni, I. (2014). Long non-coding RNAs: New players in cell differentiation and development. Nature Reviews Genetics, 15(1), 7–21.
Benetatos, L., Vartholomatos, G., & Hatzimichael, E. (2011). MEG3 imprinted gene contribution in tumorigenesis. International Journal of Cancer, 129(4), 773–779.
Wang, P., Ren, Z., & Sun, P. (2012). Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. Journal of Cellular Biochemistry, 113(6), 1868–1874.
Zhuang, W., Ge, X., Yang, S., Huang, M., Zhuang, W., Chen, P., et al. (2015). Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells From Multiple Myeloma Patients By Targeting BMP4 Transcription. Stem Cells., 33(6), 1985–1997.
Boon, R. A., Hofmann, P., Michalik, K. M., Lozano-Vidal, N., Berghauser, D., Fischer, A., et al. (2016). Long Noncoding RNA Meg3 Controls Endothelial Cell Aging and Function: Implications for Regenerative Angiogenesis. Journal of the American College of Cardiology, 68(23), 2589–2591.
Liu, X., Zhu, W., Wang, L., Wu, J., Ding, F., & Song, Y. (2019). miR-145-5p suppresses osteogenic differentiation of adipose-derived stem cells by targeting semaphorin 3A. In Vitro Cellular and Developmental Biology. Animal, 55(3), 189–202.
Liu, X., Deng, X., Ding, R., Cheng, X., & Jia, J. (2020). Chondrocyte suppression is mediated by miR-129-5p via GDF11/SMAD3 signaling in developmental dysplasia of the hip. Journal of Orthopaedic Research, 38(12), 2559–2572.
Ye, J., Lin, Y., Yu, Y., & Sun, D. (2020). LncRNA NEAT1/microRNA-129-5p/SOCS2 axis regulates liver fibrosis in alcoholic steatohepatitis. Journal of Translational Medicine, 18(1), 445.
Chen, T., Wu, Q., Zhang, Y., Lu, T., Yue, W., & Zhang, D. (2016). Tcf4 Controls Neuronal Migration of the Cerebral Cortex through Regulation of Bmp7. Frontiers in Molecular Neuroscience, 9, 94.
Wang, L., Wu, F., Song, Y., Li, X., Wu, Q., Duan, Y., et al. (2016). Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death & Disease, 7(8), e2327.
Peterson, J. R., Agarwal, S., Brownley, R. C., Loder, S. J., Ranganathan, K., Cederna, P. S., et al. (2015). Direct Mouse Trauma/Burn Model of Heterotopic Ossification. Journal of Visualized Experiments, 102, e52880.
Sakuma, C., Sato, T., Shibata, T., Nakagawa, M., Kurosawa, Y., Okumura, C. J., et al. (2021). Western blotting analysis of proteins separated by agarose native gel electrophoresis. International Journal of Biological Macromolecules, 166, 1106–1110.
Gregory, C. A., Gunn, W. G., Peister, A., & Prockop, D. J. (2004). An Alizarin red-based assay of mineralization by adherent cells in culture: Comparison with cetylpyridinium chloride extraction. Analytical Biochemistry, 329(1), 77–84.
Vaishya, R., Maduka, C. O., Agarwal, A. K., Vijay, V., & Vaish, A. (2019). Heterotopic Ossification of Tendo Achilles: An Uncommon Clinical Entity. J Orthop Case Rep., 9(2), 45–47.
Agabalyan, N. A., Evans, D. J., & Stanley, R. L. (2013). Investigating tendon mineralisation in the avian hindlimb: A model for tendon ageing, injury and disease. Journal of Anatomy, 223(3), 262–277.
Cholok, D., Chung, M. T., Ranganathan, K., Ucer, S., Day, D., Davis, T. A., et al. (2018). Heterotopic ossification and the elucidation of pathologic differentiation. Bone, 109, 12–21.
Zhang, Q., Zhou, D., Wang, H., & Tan, J. (2020). Heterotopic ossification of tendon and ligament. Journal of Cellular and Molecular Medicine, 24(10), 5428–5437.
Rui, Y. F., Lui, P. P., Li, G., Fu, S. C., Lee, Y. W., & Chan, K. M. (2010). Isolation and characterization of multipotent rat tendon-derived stem cells. Tissue Engineering Part A, 16(5), 1549–1558.
Asai, S., Otsuru, S., Candela, M. E., Cantley, L., Uchibe, K., Hofmann, T. J., et al. (2014). Tendon progenitor cells in injured tendons have strong chondrogenic potential: The CD105-negative subpopulation induces chondrogenic degeneration. Stem Cells., 32(12), 3266–3277.
Xu, K., Zhang, Z., Chen, M., Moqbel, S. A. A., He, Y., Ma, C., et al. (2020). Nesfatin-1 Promotes the Osteogenic Differentiation of Tendon-Derived Stem Cells and the Pathogenesis of Heterotopic Ossification in Rat Tendons via the mTOR Pathway. Frontiers in Cell and Development Biology, 8, 547342.
Ulitsky, I., & Bartel, D. P. (2013). lincRNAs: Genomics, evolution, and mechanisms. Cell, 154(1), 26–46.
Wang, C. G., Hu, Y. H., Su, S. L., & Zhong, D. (2020). LncRNA DANCR and miR-320a suppressed osteogenic differentiation in osteoporosis by directly inhibiting the Wnt/beta-catenin signaling pathway. Experimental & Molecular Medicine, 52(8), 1310–1325.
Ma, J., Zhang, X., Zhang, H., & Chen, H. (2020). lncRNA MEG3 Suppresses the Progression of Ankylosis Spondylitis by Regulating the Let-7i/SOST Axis. Frontiers in Molecular Biosciences, 7, 173.
Chen, S., Jia, L., Zhang, S., Zheng, Y., & Zhou, Y. (2018). DEPTOR regulates osteogenic differentiation via inhibiting MEG3-mediated activation of BMP4 signaling and is involved in osteoporosis. Stem Cell Research & Therapy, 9(1), 185.
Liu, Y., Zeng, X., Miao, J., Liu, C., Wei, F., Liu, D., et al. (2019). Upregulation of long noncoding RNA MEG3 inhibits the osteogenic differentiation of periodontal ligament cells. Journal of Cellular Physiology, 234(4), 4617–4626.
Geisler, S., & Coller, J. (2013). RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nature Reviews Molecular Cell Biology, 14(11), 699–712.
Salmena, L., Poliseno, L., Tay, Y., Kats, L., Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 146(3):353–8.
Wang, Q., Li, Y., Zhang, Y., Ma, L., Lin, L., Meng, J., et al. (2017). LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomedicine & Pharmacotherapy, 89, 1178–1186.
Wang, S., Xiong, G., Ning, R., Pan, Z., Xu, M., Zha, Z., et al. (2022). LncRNA MEG3 promotes osteogenesis of hBMSCs by regulating miR-21-5p / SOD3 axis. Acta Biochimica Polonica, 69(1), 71–77.
Zhu, Z., Zhang, X., Jiang, Y., Ruan, S., Huang, F., Zeng, H., et al. (2021). NEAT1 functions as a key mediator of BMP2 to promote osteogenic differentiation of renal interstitial fibroblasts. Epigenomics, 13(15), 1171–1186.
Alexander, M. S., Kawahara, G., Motohashi, N., Casar, J. C., Eisenberg, I., Myers, J. A., et al. (2013). MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death and Differentiation, 20(9), 1194–1208.
Formosa, A., Markert, E. K., Lena, A. M., Italiano, D., Finazzi-Agro, E., Levine, A. J., et al. (2014). MicroRNAs, miR-154, miR-299–5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485–3p, miR-495 and miR-654–3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene, 33(44):5173–82.
Chiarini, F., Paganelli, F., Martelli, A. M., & Evangelisti, C. (2020). The role played by Wnt/beta-catenin signaling pathway in acute lymphoblastic leukemia. International Journal of Molecular Sciences, 21(3), 1098.
Xu, L., Lin, W., Wen, L., & Li, G. (2019). Lgr5 in cancer biology: Functional identification of Lgr5 in cancer progression and potential opportunities for novel therapy. Stem Cell Research & Therapy, 10(1), 219.
Tu, B., Yu, B., Wang, W., Li, J., Yuan, F., Zhu, J., et al. (2021). Inhibition of IL-17 prevents the progression of traumatic heterotopic ossification. Journal of Cellular and Molecular Medicine, 25(16), 7709–7719.
Wang, Y., Rattner, A., Zhou, Y., Williams, J., Smallwood, P. M., & Nathans, J. (2012). Norrin/Frizzled4 signaling in retinal vascular development and blood brain barrier plasticity. Cell, 151(6), 1332–1344.
Bhanot, P., Brink, M., Samos, C. H., Hsieh, J. C., Wang, Y., Macke, J. P., et al. (1996). A new member of the frizzled__family from Drosophila functions as a Wingless__receptor. Nature, 382:225–30.
Pinson, K. I., Brennan, J., Monkley, S., Avery, B. J., Skarnes, W. C. (2000). An LDL-receptor-related protein mediates__Wnt signalling in mice. Nature, 407:535–8.
Acknowledgements
We appreciated the support from Base for Interdisciplinary Innovative Talent Training, Shanghai Jiao Tong University and Youth Science and Technology Innovation Studio of Shanghai Jiao Tong University School of Medicine.
Funding
This study were supported by National Natural Science Foundation of China (81830076, 8217090787); Shanghai Engineering Technology Research Center and Professional Technology Service Platform project of 2020 “Science and Technology Innovation Action Plan” of Shanghai (20DZ2254100); Municipal Hospital Clinical Skills and Innovation Capacity of Three-year Action Plan Program of Shanghai Shenkang Hospital Development Center (SHDC2020CR2039B, SHDC2020CR6019-002); Biomedical Technology Support Special Project of Shanghai “Science and Technology Innovation Action Plan” (20S31900300, 21S31902300); Clinical Research Center (CRC) of Shanghai University of Medicine and Health Sciences (20MC2020001); Research on transformation chain and transformation mode of scientific and technological achievements in public hospitals-Take Class A tertiary hospital in Shanghai as an example (lygl202214).
All the funding body have not participated in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
HL and ZYS designed and conducted the in vitro and in vivo experiments, analyzed the data and wrote the manuscript.
YHH conducted some of the in vivo experiments, micro-CT, IHC, IF and WB analysis.
GL, XW and ZJT conducted some of the in vitro experiments, ALP, ARS, IF and WB analysis.
HJR conducted some of experiments of PCR and ELISA.
BT and ZYS conducted some of experiments of animal model and cell culture.
CYF designed and conducted the research, wrote the manuscript, directed the project, and provided funding.
JHL conducted the research, wrote the manuscript.
HL, ZYS, JHL and CYF have read and verified the underlying data.
All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
All experiments and procedures were approved by the ethics committee of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital.
Consent for Publication
All authors consent to publish this manuscript.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gang Luo and Yuehao Hu contribute equally to this work as co-first authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, H., Sun, Z., Luo, G. et al. lncRNA MEG3 Promotes Osteogenic Differentiation of Tendon Stem Cells Via the miR-129-5p/TCF4/β-Catenin Axis and thus Contributes to Trauma-Induced Heterotopic Ossification. Stem Cell Rev and Rep 19, 2311–2328 (2023). https://doi.org/10.1007/s12015-023-10562-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-023-10562-w