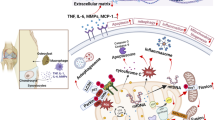
Abstract
During the last two decades, mesenchymal stem cells (MSCs) gained a place of privilege in the field of regenerative medicine. Recently, extracellular vesicles (EVs) have been identified as major mediators of MSCs immunosuppressive as well as pro-regenerative activities in many disease models, including inflammatory/degenerative conditions as joint diseases and osteoarthritis. In order to shed light on EVs potential, a rigorous profiling of embedded proteins, lipids and nucleic acids (mRNA/miRNA) is mandatory. Nevertheless, reliable strategies to efficiently score miRNA cargo and modulation under diverse experimental conditions or treatments are missing. The aim of this work was to identify reliable reference genes (RGs) to analyze miRNA content in EVs secreted by adipose-derived MSCs (ASCs) and verify their consistency under inflammatory conditions that were proposed to enhance ASC-EVs immunomodulatory and regenerative potential. RefFinder algorithm, that integrates the currently available major computational programs (geNorm, NormFinder, BestKeeper, and Delta Ct method), allowed to identify miR-22-5p and miR-29a-5p as the most stable RGs. Notably, both miRNAs maintained the highest stability when EVs isolated from IFNg-treated ASCs were included in the analysis. In addition, considerable effects of suboptimal RGs choice on the reliable quantification of miRNAs involved at different levels (tissue homeostasis or macrophage polarization) in the osteoarthritis phenotype, and thus considered as promising therapeutic molecule, have clearly been demonstrated. In conclusion, a proper normalization method is not only needed for research purposes but also mandatory to characterize clinical products and predict their therapeutic potential, especially in the emerging field of MSCs derived-EVs as new tools for regenerative medicine.




Similar content being viewed by others
References
Law, S., & Chaudhuri, S. (2013). Mesenchymal stem cell and regenerative medicine: Regeneration versus immunomodulatory challenges. American Journal of Stem Cells, 2(1), 22–38.
Klinker, M. W., & Wei, C. H. (2015). Mesenchymal stem cells in the treatment of inflammatory and autoimmune diseases in experimental animal models. World Journal of Stem Cells, 7, 556–567. https://doi.org/10.4252/wjsc.v7.i3.556.
Puissant, B., Barreau, C., Bourin, P., Clavel, C., Corre, J., Bousquet, C., Taureau, C., Cousin, B., Abbal, M., Laharrague, P., Penicaud, L., Casteilla, L., & Blancher, A. (2005). Immunomodulatory effect of human adipose tissue-derived adult stem cells: Comparison with bone marrow mesenchymal stem cells. British Journal of Haematology, 129, 118–129. https://doi.org/10.1111/j.1365-2141.2005.05409.x.
Hoogduijn, M. J., Crop, M. J., Peeters, A. M., et al. (2007). Human heart, spleen, and perirenal fat-derived mesenchymal stem cells have immunomodulatory capacities. Stem Cells and Development, 16, 597–604. https://doi.org/10.1089/scd.2006.0110.
Yañez, R., Lamana, M. L., García-Castro, J., Colmenero, I., Ramírez, M., & Bueren, J. A. (2006). Adipose tissue-derived mesenchymal stem cells have in vivo immunosuppressive properties applicable for the control of the graft-versus-host disease. Stem Cells, 24, 2582–2591. https://doi.org/10.1634/stemcells.2006-0228.
González, M. A., Gonzalez-Rey, E., Rico, L., Büscher, D., & Delgado, M. (2009). Adipose-derived mesenchymal stem cells alleviate experimental colitis by inhibiting inflammatory and autoimmune responses. Gastroenterology, 136, 978–989. https://doi.org/10.1053/j.gastro.2008.11.041.
Crop, M. J., Baan, C. C., Korevaar, S. S., IJzermans, J. N. M., Pescatori, M., Stubbs, A. P., van IJcken, W. F. J., Dahlke, M. H., Eggenhofer, E., Weimar, W., & Hoogduijn, M. J. (2010). Inflammatory conditions affect gene expression and function of human adipose tissue-derived mesenchymal stem cells. Clinical and Experimental Immunology, 162, 474–486. https://doi.org/10.1111/j.1365-2249.2010.04256.x.
Manfredini, C., Maumus, M., Gabusi, E., et al. (2013). Adipose-derived mesenchymal stem cells exert antiinflammatory effects on chondrocytes and synoviocytes from osteoarthritis patients through prostaglandin E2. Arthritis and Rheumatism, 65, 1271–1281. https://doi.org/10.1002/art.37908.
Veronesi, F., Maglio, M., Tschon, M., Aldini, N. N., & Fini, M. (2014). Adipose-derived mesenchymal stem cells for cartilage tissue engineering: State-of-the-art in in vivo studies. Journal of Biomedical Materials Research. Part A, 102, 2448–2466. https://doi.org/10.1002/jbm.a.34896.
van Buul, G. M., Villafuertes, E., Bos, P. K., Waarsing, J. H., Kops, N., Narcisi, R., Weinans, H., Verhaar, J. A. N., Bernsen, M. R., & van Osch, G. J. V. M. (2012). Mesenchymal stem cells secrete factors that inhibit inflammatory processes in short-term osteoarthritic synovium and cartilage explant culture. Osteoarthritis and Cartilage, 20, 1186–1196. https://doi.org/10.1016/j.joca.2012.06.003.
Montemurro, T., Viganò, M., Ragni, E., Barilani, M., Parazzi, V., Boldrin, V., Lavazza, C., Montelatici, E., Banfi, F., Lauri, E., Giovanelli, S., Baccarin, M., Guerneri, S., Giordano, R., & Lazzari, L. (2016). Angiogenic and anti-inflammatory properties of mesenchymal stem cells from cord blood: Soluble factors and extracellular vesicles for cell regeneration. European Journal of Cell Biology, 95, 228–238. https://doi.org/10.1016/j.ejcb.2016.04.003.
Tofiño-Vian, M., Guillén, M. I., & Alcaraz, M. J. (2018). Extracellular vesicles: A new therapeutic strategy for joint conditions. Biochemical Pharmacology, 153, 134–146. https://doi.org/10.1016/j.bcp.2018.02.004.
Tofiño-Vian, M., Guillén, M. I., Pérez Del Caz, M. D., Silvestre, A., & Alcaraz, M. J. (2018). Microvesicles from human adipose tissue-derived mesenchymal stem cells as a new protective strategy in osteoarthritic chondrocytes. Cellular Physiology and Biochemistry, 47, 11–25. https://doi.org/10.1159/000489739.
Cosenza, S., Ruiz, M., Toupet, K., Jorgensen, C., & Noël, D. (2017). Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Scientific Reports, 7, 16214. https://doi.org/10.1038/s41598-017-15376-8.
Blazquez, R., Sanchez-Margallo, F. M., de la Rosa, O., Dalemans, W., Alvarez, V., Tarazona, R., & Casado, J. G. (2014). Immunomodulatory potential of human adipose mesenchymal stem cells derived exosomes on in vitro stimulated T cells. Frontiers in Immunology, 5. https://doi.org/10.3389/fimmu.2014.00556.
van Niel, G., D'Angelo, G., & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews. Molecular Cell Biology, 19, 213–228. https://doi.org/10.1038/nrm.2017.125.
Yáñez-Mó, M., Siljander, P. R., Andreu, Z., et al. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4. https://doi.org/10.3402/jev.v4.27066.
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297.
Ti, D., Hao, H., Fu, X., & Han, W. (2016). Mesenchymal stem cells-derived exosomal microRNAs contribute to wound inflammation. Science China. Life Sciences, 59, 1305–1312. https://doi.org/10.1007/s11427-016-0240-4.
Li, X., Liu, L., Yang, J., Yu, Y., Chai, J., Wang, L., Ma, L., & Yin, H. (2016). Exosome derived from human umbilical cord mesenchymal stem cell mediates MiR-181c attenuating burn-induced excessive inflammation. EBioMedicine, 8, 72–82. https://doi.org/10.1016/j.ebiom.2016.04.030.
Lo Sicco, C., Reverberi, D., Balbi, C., Ulivi, V., Principi, E., Pascucci, L., Becherini, P., Bosco, M. C., Varesio, L., Franzin, C., Pozzobon, M., Cancedda, R., & Tasso, R. (2017). Mesenchymal stem cell-derived extracellular vesicles as mediators of anti-inflammatory effects: Endorsement of macrophage polarization. Stem Cells Translational Medicine, 6, 1018–1028. https://doi.org/10.1002/sctm.16-0363.
Redondo-Castro, E., Cunningham, C., Miller, J., Martuscelli, L., Aoulad-Ali, S., Rothwell, N. J., Kielty, C. M., Allan, S. M., & Pinteaux, E. (2017). Interleukin-1 primes human mesenchymal stem cells towards an anti-inflammatory and pro-trophic phenotype in vitro. Stem Cell Research & Therapy, 8, 79. https://doi.org/10.1186/s13287-017-0531-4.
Ti, D., Hao, H., Tong, C., Liu, J., Dong, L., Zheng, J., Zhao, Y., Liu, H., Fu, X., & Han, W. (2015). LPS-preconditioned mesenchymal stromal cells modify macrophage polarization for resolution of chronic inflammation via exosome-shuttled let-7b. Journal of Translational Medicine, 13, 308. https://doi.org/10.1186/s12967-015-0642-6.
Domenis, R., Cifù, A., Quaglia, S., Pistis, C., Moretti, M., Vicario, A., Parodi, P. C., Fabris, M., Niazi, K. R., Soon-Shiong, P., & Curcio, F. (2018). Pro inflammatory stimuli enhance the immunosuppressive functions of adipose mesenchymal stem cells-derived exosomes. Scientific Reports, 8, 13325. https://doi.org/10.1038/s41598-018-31707-9.
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., Mueller, R., Nolan, T., Pfaffl, M. W., Shipley, G. L., Vandesompele, J., & Wittwer, C. T. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55, 611–622. https://doi.org/10.1373/clinchem.2008.112797.
Li, Y., Xiang, G. M., Liu, L. L., et al. (2015). Assessment of endogenous reference gene suitability for serum exosomal microRNA expression analysis in liver carcinoma resection studies. Molecular Medicine Reports, 12, 4683–4691. https://doi.org/10.3892/mmr.2015.3919.
Li, Y., Zhang, L., Liu, F., Xiang, G., Jiang, D., & Pu, X. (2015). Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Disease Markers, 2015, 1–12. https://doi.org/10.1155/2015/893594.
Garcia-Contreras, M., Shah, S. H., Tamayo, A., Robbins, P. D., Golberg, R. B., Mendez, A. J., & Ricordi, C. (2017). Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration type 1 diabetes. Scientific Reports, 7, 5998. https://doi.org/10.1038/s41598-017-05787-y.
Santovito, D., De Nardis, V., Marcantonio, P., et al. (2014). Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: Effect of glycemic control. The Journal of Clinical Endocrinology and Metabolism, 99, E1681–E1685. https://doi.org/10.1210/jc.2013-3843.
Gouin, K., Peck, K., Antes, T., Johnson, J. L., Li, C., Vaturi, S. D., Middleton, R., de Couto, G., Walravens, A. S., Rodriguez-Borlado, L., Smith, R. R., Marbán, L., Marbán, E., & Ibrahim, A. G. E. (2017). A comprehensive method for identification of suitable reference genes in extracellular vesicles. Journal of Extracellular Vesicles, 6. https://doi.org/10.1080/20013078.2017.1347019.
Benz, F., Roderburg, C., Vargas Cardenas, D., Vucur, M., Gautheron, J., Koch, A., Zimmermann, H., Janssen, J., Nieuwenhuijsen, L., Luedde, M., Frey, N., Tacke, F., Trautwein, C., & Luedde, T. (2013). U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Experimental & Molecular Medicine, 45, e42. https://doi.org/10.1038/emm.2013.81.
Johnson, C. I., Argyle, D. J., & Clements, D. N. (2016). In vitro models for the study of osteoarthritis. Veterinary Journal, 209, 40–49. https://doi.org/10.1016/j.tvjl.2015.07.011.
Lopa, S., Colombini, A., Stanco, D., de Girolamo, L., Sansone, V., & Moretti, M. (2014). Donor-matched mesenchymal stem cells from knee infrapatellar and subcutaneous adipose tissue of osteoarthritic donors display differential chondrogenic and osteogenic commitment. European Cells & Materials, 27, 298–311.
Ragni, E., Montemurro, T., Montelatici, E., Lavazza, C., Viganò, M., Rebulla, P., Giordano, R., & Lazzari, L. (2013). Differential microRNA signature of human mesenchymal stem cells from different sources reveals an "environmental-niche memory" for bone marrow stem cells. Experimental Cell Research, 319, 1562–1574. https://doi.org/10.1016/j.yexcr.2013.04.002.
Müller, G. (2012). Novel tools for the study of cell type-specific exosomes and microvesicles. Journal of Bioanalysis and Biomedicine, 04. https://doi.org/10.4172/1948-593X.1000063.
Cavalleri, T., Angelici, L., Favero, C., Dioni, L., Mensi, C., Bareggi, C., Palleschi, A., Rimessi, A., Consonni, D., Bordini, L., Todaro, A., Bollati, V., & Pesatori, A. C. (2017). Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS One, 12, e0176680. https://doi.org/10.1371/journal.pone.0176680.
Metsalu, T., & Vilo, J. (2015). ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Research, 43, W566–W570. https://doi.org/10.1093/nar/gkv468.
Andersen, C. L., Jensen, J. L., & Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496.
Vandesompele, J., De Preter, K., Pattyn, F., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7) RESEARCH0034.
Pfaffl, M. W., Tichopad, A., Prgomet, C., & Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--excel-based tool using pair-wise correlations. Biotechnology Letters, 26(6), 509–515.
Silver, N., Best, S., Jiang, J., & Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology, 7, 33. https://doi.org/10.1186/1471-2199-7-33.
Xie, F., Xiao, P., Chen, D., Xu, L., & Zhang, B. (2012). miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Molecular Biology, 80, 75–84. https://doi.org/10.1007/s11103-012-9885-2.
Ramos, L. T., Sánchez-Abarca, L. I., Muntión, S., et al. (2016). MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Communication and Signaling: CCS, 14, 2. https://doi.org/10.1186/s12964-015-0124-8.
Shi, D. L., Shi, G. R., Xie, J., Du, X. Z., & Yang, H. (2016). MicroRNA-27a inhibits cell migration and invasion of fibroblast-like Synoviocytes by targeting Follistatin-like protein 1 in rheumatoid arthritis. Molecules and Cells, 39, 611–618. https://doi.org/10.14348/molcells.2016.0103.
Zhang, M., Lygrisse, K., & Wang, J. (2017). Role of MicroRNA in osteoarthritis. Journal of Arthritis, 06. https://doi.org/10.4172/2167-7921.1000239.
Essandoh, K., Li, Y., Huo, J., & Fan, G. C. (2016). MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock, 46, 122–131. https://doi.org/10.1097/SHK.0000000000000604.
Miana, V. V., & González, E. A. P. (2018). Adipose tissue stem cells in regenerative medicine. Ecancermedicalscience, 12. https://doi.org/10.3332/ecancer.2018.822.
Palombella, S., Pirrone, C., Cherubino, M., Valdatta, L., Bernardini, G., & Gornati, R. (2017). Identification of reference genes for qPCR analysis during hASC long culture maintenance. PLoS One, 12, e0170918. https://doi.org/10.1371/journal.pone.0170918.
Schildberg, T., Rauh, J., Bretschneider, H., & Stiehler, M. (2013). Identification of suitable reference genes in bone marrow stromal cells from osteoarthritic donors. Stem Cell Research, 11, 1288–1298. https://doi.org/10.1016/j.scr.2013.08.015.
Ragni, E., Viganò, M., Rebulla, P., Giordano, R., & Lazzari, L. (2013). What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: How to choose the most reliable housekeeping genes. Journal of Cellular and Molecular Medicine, 17, 168–180. https://doi.org/10.1111/j.1582-4934.2012.01660.x.
Banfi, F., Colombini, A., Perucca Orfei, C., Parazzi, V., & Ragni, E. (2018). Validation of reference and identity-defining genes in human mesenchymal stem cells cultured under unrelated fetal bovine serum batches for basic science and clinical application. Stem Cell Reviews, 14, 837–846. https://doi.org/10.1007/s12015-018-9822-0.
Viganò, M., Perucca Orfei, C., de Girolamo, L., Pearson, J. R., Ragni, E., de Luca, P., & Colombini, A. (2018). Housekeeping gene stability in human mesenchymal stem and tendon cells exposed to Tenogenic factors. Tissue Engineering. Part C, Methods, 24, 360–367. https://doi.org/10.1089/ten.TEC.2017.0518.
Schwarzenbach, H., da Silva, A. M., Calin, G., & Pantel, K. (2015). Data normalization strategies for MicroRNA quantification. Clinical Chemistry, 61, 1333–1342. https://doi.org/10.1373/clinchem.2015.239459.
Didychuk, A. L., Butcher, S. E., & Brow, D. A. (2018). The life of U6 small nuclear RNA, from cradle to grave. RNA, 24, 437–460. https://doi.org/10.1261/rna.065136.117.
Terns, M. P., Dahlberg, J. E., & Lund, E. (1993). Multiple cis-acting signals for export of pre-U1 snRNA from the nucleus. Genes & Development, 7(10), 1898–1908.
Shumyatsky, G., Wright, D., & Reddy, R. (1993). Methylphosphate cap structure increases the stability of 7SK, B2 and U6 small RNAs in Xenopus oocytes. Nucleic Acids Research, 21(20), 4756–4761.
Lv, C., & Yang, T. (2018). Effective enrichment of urinary exosomes by polyethylene glycol for RNA detection. Biomedical Research, 29. https://doi.org/10.4066/biomedicalresearch.29-17-1695.
Xu, J. F., Wang, Y. P., Zhang, S. J., Chen, Y., Gu, H. F., Dou, X. F., Xia, B., Bi, Q., & Fan, S. W. (2017). Exosomes containing differential expression of microRNA and mRNA in osteosarcoma that can predict response to chemotherapy. Oncotarget, 8, 75968–75978. https://doi.org/10.18632/oncotarget.18373.
Hayashi, T., Lombaert, I. M., Hauser, B. R., Patel, V. N., & Hoffman, M. P. (2017). Exosomal MicroRNA transport from salivary mesenchyme regulates epithelial progenitor expansion during organogenesis. Developmental Cell, 40, 95–103. https://doi.org/10.1016/j.devcel.2016.12.001.
Zhao, L., Yu, J., Wang, J., Li, H., Che, J., & Cao, B. (2017). Isolation and identification of miRNAs in exosomes derived from serum of colon cancer patients. Journal of Cancer, 8, 1145–1152. https://doi.org/10.7150/jca.18026.
Zhang, W., Ni, M., Su, Y., Wang, H., Zhu, S., Zhao, A., & Li, G. (2018). MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. European Urology Focus, 4, 412–419. https://doi.org/10.1016/j.euf.2016.09.007.
Occhipinti, G., Giulietti, M., Principato, G., & Piva, F. (2016). The choice of endogenous controls in exosomal microRNA assessments from biofluids. Tumour Biology, 37, 11657–11665. https://doi.org/10.1007/s13277-016-5164-1.
Will, C. L., & Lührmann, R. (2011). Spliceosome structure and function. Cold Spring Harbor Perspectives in Biology, 3. https://doi.org/10.1101/cshperspect.a003707.
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Rådmark, O., Kim, S., & Kim, V. N. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature, 425, 415–419. https://doi.org/10.1038/nature01957.
Meng, X., Xue, M., Xu, P., Hu, F., Sun, B., & Xiao, Z. (2017). MicroRNA profiling analysis revealed different cellular senescence mechanisms in human mesenchymal stem cells derived from different origin. Genomics, 109, 147–157. https://doi.org/10.1016/j.ygeno.2017.02.003.
Wang, X., Zhu, Y., Xu, B., Wang, J., & Liu, X. (2016). Identification of TLR2 and TLR4-induced microRNAs in human mesenchymal stem cells and their possible roles in regulating TLR signals. Molecular Medicine Reports, 13, 4969–4980. https://doi.org/10.3892/mmr.2016.5197.
Liu, B., Zhang, M., Zhao, J., Zheng, M., & Yang, H. (2018). Imbalance of M1/M2 macrophages is linked to severity level of knee osteoarthritis. Experimental and Therapeutic Medicine. https://doi.org/10.3892/etm.2018.6852.
Acknowledgements
Authors thank Laura Dioni, Laura Cantone, Laura Perego and Mirjam Hoxha for their help and useful discussions. The present work was funded by Ministero della Salute, Ricerca Corrente.
Author information
Authors and Affiliations
Contributions
Enrico Ragni: collection and/or assembly of data, data analysis and interpretation, conception and design, manuscript writing; Carlotta Perucca Orfei: collection and/or assembly of data, data analysis and interpretation; Paola De Luca: collection and/or assembly of data; Marco Viganò: data analysis and interpretation; Alessandra Colombini: data analysis and interpretation; Gaia Lugano: collection and/or assembly of data; Valentina Bollati: data analysis and interpretation; Laura de Girolamo: conception and design, manuscript writing, final approval of manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no conflict of interest.
Ethical Approval and Informed Consent
The study was carried out at IRCCS Istituto Ortopedico Galeazzi with Institutional Review Board approval and specimens were collected with patient informed consent (M-SPER-015 - Ver. 2–04.11.2016), and under the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 34 kb)
Rights and permissions
About this article
Cite this article
Ragni, E., Perucca Orfei, C., De Luca, P. et al. miR-22-5p and miR-29a-5p Are Reliable Reference Genes for Analyzing Extracellular Vesicle-Associated miRNAs in Adipose-Derived Mesenchymal Stem Cells and Are Stable under Inflammatory Priming Mimicking Osteoarthritis Condition. Stem Cell Rev and Rep 15, 743–754 (2019). https://doi.org/10.1007/s12015-019-09899-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-019-09899-y