Abstract
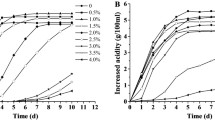
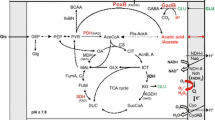
Acetic acid bacteria have a remarkable capacity to cope with elevated concentrations of cytotoxic acetic acid in their fermentation environment. In particular, the high-level acetate tolerance of Acetobacter pasteurianus that occurs in vinegar industrial settings must be constantly selected for. However, the improved acetic acid tolerance is rapidly lost without a selection pressure. To understand genetic and molecular biology of this acquired acetic acid tolerance in A. pasteurianus, we evolved three strains A. pasteurianus CICIM B7003, CICIM B7003-02, and ATCC 33,445 over 960 generations (4 months) in two initial acetic acids of 20 g·L−1 and 30 g·L−1, respectively. An acetic acid-adapted strain M20 with significantly improved specific growth rate of 0.159 h−1 and acid productivity of 1.61 g·L−1·h−1 was obtained. Comparative genome analysis of six evolved strains revealed that the genetic variations of adaptation were mainly focused on lactate metabolism, membrane proteins, transcriptional regulators, transposases, replication, and repair system. Among of these, lactate dehydrogenase, acetolactate synthase, glycosyltransferase, ABC transporter ATP-binding protein, two-component regulatory systems, the type II toxin-antitoxin system (RelE/RelB/StbE), exodeoxyribonuclease III, type I restriction endonuclease, tRNA-uridine 2-sulfurtransferase, and transposase might collaboratively contribute to the improved acetic acid tolerance in A. pasteurianus strains. The balance between repair factors and transposition variations might be the basis for genomic plasticity of A. pasteurianus strains, allowing the survival of populations and their offspring in acetic acid stress fluctuations. These observations provide important insights into the nature of acquired acetic acid tolerance phenotype and lay a foundation for future genetic manipulation of these strains.





Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its Supplementary material.
References
Lynch, K. M., Zannini, E., Wilkinson, S., Daenen, L., & Arendt, E. K. (2019). Physiology of acetic acid bacteria and their role in vinegar and fermented beverages. Comprehensive Reviews in Food Science and Food, 18, 587–625.
Yang, H., Chen, T., Wang, M., Zhou, J., Liebl, W., Barja, F., & Chen, F. (2022). Molecular biology: Fantastic toolkits to improve knowledge and application of acetic acid bacteria. Biotechnology Advances, 58, 107911.
Steiner, P., & Sauer, U. (2003). Long-term continuous evolution of acetate resistant Acetobacter aceti. Biotechnology and Bioengineering, 84, 40–44.
Qi, Z., Yang, H., Xia, X., Quan, W., Wang, W., & Yu, X. (2014). Achieving high strength vinegar fermentation via regulating cellular growth status and aeration strategy. Process Biochemistry, 49, 1063–1070.
Zorraquino, V., Kim, M., Rai, N., & Tagkopoulos, I. (2016). The genetic and transcriptional basis of short and long term adaptation across multiple stresses in Escherichia coli. Molecular Biology and Evolution, 34, 707–717.
Swings, T., Weytjens, B., Schalck, T., Bonte, C., Verstraeten, N., Michiels, J., & Marchal, K. (2017). Network-based identification of adaptive pathways in evolved ethanol-tolerant bacterial populations. Molecular Biology and Evolution, 228, 2927–2943.
Qian, C., Ma, J., Liang, J., Zhang, L., & Liang, X. (2022). Comprehensive deciphering prophages in genus Acetobacter on the ecology, genomic features, toxin-antitoxin system, and linkage with CRISPR-Cas system. Frontiers in Microbiology, 13, 951030.
Matsutani, M., Hirakawa, H., Hiraoka, E., Theeragool, G., Yakushi, T., & Matsushita, K. (2016). Complete genome sequencing and comparative genomic analysis of the thermotolerant acetic acid bacterium, Acetobacter pasteurianus SKU1108, provide a new insight into thermotolerance. Microbes and Environments, 31, 395–400.
Casacuberta, E., & González, J. (2013). The impact of transposable elements in environmental adaptation. Molecular Ecology, 22, 1503–1517.
Gullo, M., Verzelloni, E., & Canonico, M. (2014). Aerobic submerged fermentation by acetic acid bacteria for vinegar production: Process and biotechnological aspects. Process Biochemistry, 49, 1571–1579.
Gao, L., Wu, X., Zhu, C., Jin, Z., Wang, W., & Xia, X. (2020). Metabolic engineering to improve the biomanufacturing efficiency of acetic acid bacteria: Advances and prospects. Critical Reviews in Biotechnology, 40, 522–538.
Yang, H., Yu, Y., Fu, C., & Chen, F. (2019). Bacterial acid resistance toward organic weak acid revealed by RNA-Seq transcriptomic analysis in Acetobacter pasteurianus. Frontiers in Microbiology, 10, 1616.
Qi, Z., Wang, W., Yang, H., Xia, X., & Yu, X. (2014). Mutation of Acetobacter pasteurianus by UV irradiation under acidic stress for high-acidity vinegar fermentation. International Journal of Food Science & Technology, 49, 468–476.
Xia, X., Zhu, X., Yang, H., Xin, Y., & Wang, W. (2015). Enhancement of rice vinegar production by modified semi-continuous culture based on analysis of enzymatic kinetic. European Food Research and Technology, 241, 479–485.
Qi, Z., Yang, H., Xia, X., Wang, W., & Yu, X. (2014). High strength vinegar fermentation by acetobacter pasteurianus via enhancing alcohol respiratory chain. Biotechnology and Bioprocess Engineering, 19, 289–297.
Quinlan, A. R. (2014). BEDTools: The Swiss-Army tool for genome feature analysis. Current Protocols in Bioinformatics 47, 11.12.1-11.12.34.
Wang, B., Shao, Y., Chen, T., Chen, W., & Chen, F. (2015). Global insights into acetic acid resistance mechanisms and genetic stability of Acetobacter pasteurianus strains by comparative genomics. Scientific Reports-Uk, 5, 18330.
Sato, J., Wakayama, M., & Takagi, K. (2015). Lactate dehydrogenase involved in lactate metabolism of Acetobacter pasteurianus. Procedia Environmental Sciences, 28, 67–71.
Xia, K., Zang, N., Zhang, J., Zhang, H., Li, Y., Liu, Y., Feng, W., & Liang, X. (2016). New insights into the mechanisms of acetic acid resistance in Acetobacter pasteurianus using iTRAQ-dependent quantitative proteomic analysis. International Journal of Food Microbiology, 238, 241–251.
Christel, S., Fridlund, J., Watkin, E. L., & Dopson, M. (2016). Acidithiobacillus ferrivorans SS3 presents little RNA transcript response related to cold stress during growth at 8 °C suggesting it is a eurypsychrophile. Extremophiles, 20, 903–913.
Matsumoto, N., Hattori, H., Matsutani, M., Matayoshi, C., Toyama, H., Kataoka, N., Yakushi, T., & Matsushita, K. (2018). A single-nucleotide insertion in a drug transporter gene induces a thermotolerant phenotype of Gluconobacter frateurii by increasing the NADPH/NADP+ ratio via metabolic change. Applied and Environment Microbiology, 84, e00354.
Zhu, Z., Zhang, J., Ji, X., Fang, Z., Wu, Z., Chen, J., & Du, G. (2018). Evolutionary engineering of industrial microorganisms-strategies and applications. Applied Microbiology and Biotechnology, 102, 4615–4627.
Qiu, X., Zhang, Y., & Hong, H. (2021). Classification of acetic acid bacteria and their acid resistant mechanism. AMB Express, 11, 29.
Zheng, Y., Zhang, K. P., Su, G. Y., Han, Q., Shen, Y. B., & Wang, M. (2015). The evolutionary response of alcohol dehydrogenase and aldehyde dehydrogenases of Acetobacter pasteurianus CGMCC 3089 to ethanol adaptation. Food Science and Biotechnology, 24, 133–140.
Monedero, V., Revilla-Guarinos, A., & Zúñiga, M. (2017). Physiological role of two-component signal transduction systems in food-associated lactic acid bacteria. Advances in Applied Microbiology, 99, 1–51.
Hempel, N., Görisch, H., & Mern, D. S. (2013). Gene ercA, encoding a putative iron-containing alcohol dehydrogenase, is involved in regulation of ethanol utilization in Pseudomonas aeruginosa. Journal of Bacteriology, 195, 3925–3932.
Bockwoldt, J. A., Meng, C., Ludwig, C., Kupetz, M., & Ehrmann, M. A. (2022). Proteomic analysis reveals enzymes for β-D-glucan formation and degradation in Levilactobacillus brevis TMW 1.2112. International Journal of Molecular Sciences, 23, 3393.
Brandt, J. U., Born, F.-L., Jakob, F., & Vogel, R. F. (2017). Environmentally triggered genomic plasticity and capsular polysaccharide formation are involved in increased ethanol and acetic acid tolerance in Kozakia baliensis NBRC 16680. BMC Microbiology, 17, 172.
Hu, Y., Hu, Q., Wei, R., Li, R., Zhao, D., Ge, M., Yao, Q., & Yu, X. (2018). The XRE family transcriptional regulator SrtR in Streptococcus suis is involved in oxidant tolerance and virulence. Frontiers in Cellular and Infection Microbiology, 8, 452.
Zhu, Z., Yang, J., Yang, P., Wu, Z., Zhang, J., & Du, G. (2019). Enhanced acid-stress tolerance in Lactococcus lactis NZ9000 by overexpression of ABC transporters. Microbial Cell Factories, 18, 136.
Souza, L. L., Eduardo, I. R., Pádula, M., & Leitão, A. C. (2006). Endonuclease IV and exonuclease III are involved in the repair and mutagenesis of DNA lesions induced by UVB in Escherichia coli. Mutagenesis, 21, 125–130.
Kaneda, K., Sekiguchi, J., & Shida, T. (2006). Role of the tryptophan residue in the vicinity of the catalytic center of exonuclease III family AP endonucleases: AP site recognition mechanism. Nucleic Acids Research, 34, 1552–1563.
Black, K. A., & Dos Santos, P. C. (2015). Abbreviated pathway for biosynthesis of 2-thiouridine in Bacillus subtilis. Journal of Bacteriology, 197, 1952–1962.
Somayaji, A., Dhanjal, C. R., Lingamsetty, R., Vinayagam, R., Selvaraj, R., Varadavenkatesan, T., & Govarthanan, M. (2022). An insight into the mechanisms of homeostasis in extremophiles. Microbiological Research, 263, 127115.
Funding
This work is financially supported by the National Natural Science Foundation of China (grant nos. 31901671) and the Fundamental Research Funds for the Central Universities (JUSRP121016).
Author information
Authors and Affiliations
Contributions
L. Gao and X.L. Xia conceived and designed the experiments. L. Gao and W. Shi carried out the experiments and analyzed the data. L. Gao and X.L. Xia wrote and revised the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors agreed to publish this article.
Human and Animal Participants
No such procedures were performed in studies.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, L., Shi, W. & Xia, X. Genomic Plasticity of Acid-Tolerant Phenotypic Evolution in Acetobacter pasteurianus. Appl Biochem Biotechnol 195, 6003–6019 (2023). https://doi.org/10.1007/s12010-023-04353-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04353-9