Abstract
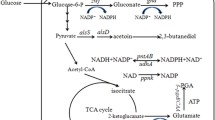
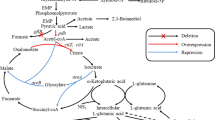
Poly-γ-glutamic acid is a multi-functional biopolymer with various applications. ATP supply plays an important role in poly-γ-glutamic acid (γ-PGA) synthesis. Global anaerobic regulator Fnr plays a key role in anaerobic adaptation and nitrate respiration, which might affect ATP generation during γ-PGA synthesis. In this study, we have improved γ-PGA production by overexpression of Fnr in Bacillus licheniformis WX-02. First, the gene fnr was knocked out in WX-02, and the γ-PGA yields have no significant differences between WX-02 and the fnr-deficient strain WXΔfnr in the medium without nitrate (BFC medium). However, the γ-PGA yield of 8.95 g/L, which was produced by WXΔfnr in the medium with nitrate addition (BFCN medium), decreased by 74% compared to WX-02 (34.53 g/L). Then, the fnr complementation strain WXΔfnr/pHY-fnr restored the γ-PGA synthesis capability, and γ-PGA yield was increased by 13% in the Fnr overexpression strain WX/pHY-fnr (39.96 g/L) in BFCN medium, compared to WX/pHY300 (35.41 g/L). Furthermore, the transcriptional levels of narK, narG, and hmp were increased by 5.41-, 4.93-, and 3.93-fold in WX/pHY-fnr, respectively, which led to the increases of nitrate consumption rate and ATP supply for γ-PGA synthesis. Collectively, Fnr affects γ-PGA synthesis mainly through manipulating the expression level of nitrate metabolism, and this study provides a novel strategy to improve γ-PGA production by overexpression of Fnr.





Similar content being viewed by others
References
Candela, T., & Fouet, A. (2006). Poly-gamma-glutamate in bacteria. Molecular Microbiology, 60(5), 1091–1098. https://doi.org/10.1111/j.1365-2958.2006.05179.x.
Feng, J., Gu, Y., Quan, Y., Cao, M., Gao, W., Zhang, W., Wang, S., Yang, C., & Song, C. (2015). Improved poly-gamma-glutamic acid production in Bacillus amyloliquefaciens by modular pathway engineering. Metabolic Engineering, 32, 106–115. https://doi.org/10.1016/j.ymben.2015.09.011.
Luo, Z., Guo, Y., Liu, J., Qiu, H., Zhao, M., Zou, W., & Li, S. (2016). Microbial synthesis of poly-gamma-glutamic acid: current progress, challenges, and future perspectives. Biotechnology for Biofuels, 9(1), 134. https://doi.org/10.1186/s13068-016-0537-7.
Li, X., Gou, X., Long, D., Ji, Z., Hu, L., Xu, D., Liu, J., & Chen, S. (2014). Physiological and metabolic analysis of nitrate reduction on poly-gamma-glutamic acid synthesis in Bacillus licheniformis WX-02. Archives of Microbiology, 196(11), 791–799. https://doi.org/10.1007/s00203-014-1014-y.
Su, Y., Li, X., Liu, Q., Hou, Z., Zhu, X., Guo, X., & Ling, P. (2010). Improved poly-gamma-glutamic acid production by chromosomal integration of the Vitreoscilla hemoglobin gene (vgb) in Bacillus subtilis. Bioresource Technology, 101(12), 4733–4736. https://doi.org/10.1016/j.biortech.2010.01.128.
Wu, G., Yan, Q., Jones, J. A., Tang, Y. J., Fong, S. S., & Koffas, M. A. G. (2016). Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends in Biotechnology, 34(8), 652–664. https://doi.org/10.1016/j.tibtech.2016.02.010.
Zhou, J., Liu, L., Shi, Z., Du, G., & Chen, J. (2009). ATP in current biotechnology: regulation, applications and perspectives. Biotechnology Advances, 27(1), 94–101. https://doi.org/10.1016/j.biotechadv.2008.10.005.
Geng, H., Zhu, Y., Mullen, K., Zuber, C. S., & Nakano, M. M. (2007). Characterization of ResDE-dependent fnr transcription in Bacillus subtilis. Journal of Bacteriology, 189(5), 1745–1755. https://doi.org/10.1128/JB.01502-06.
Nakano, M. M., & Hulett, F. M. (1997). Adaptation of Bacillus subtilis to oxygen limitation. FEMS Microbiology Letters, 157(1), 1–7. https://doi.org/10.1111/j.1574-6968.1997.tb12744.x.
Volbeda, A., Darnault, C., Renoux, O., Nicolet, Y., & Fontecilla-Camps, J. C. (2015). The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium. Science Advances, 1(11), e1501086. https://doi.org/10.1126/sciadv.1501086.
Marzan, L. W., Siddiquee, K. A., & Shimizu, K. (2011). Metabolic regulation of an fnr gene knockout Escherichia coli under oxygen limitation. Bioeng Bugs, 2(6), 331–337. https://doi.org/10.4161/bbug.2.6.16350.
Shan, Y., Pan, Q., Liu, J., Huang, F., Sun, H., Nishino, K., & Yan, A. (2012). Covalently linking the Escherichia coli global anaerobic regulator FNR in tandem allows it to function as an oxygen stable dimer. Biochem Biophy Res Commun, 419(1), 43–48. https://doi.org/10.1016/j.bbrc.2012.01.121.
Hartig, E., & Jahn, D. (2012). Regulation of the anaerobic metabolism in Bacillus subtilis. Advances in Microbial Physiology, 61, 195–216. https://doi.org/10.1016/B978-0-12-394423-8.00005-6.
Reents, H., Munch, R., Dammeyer, T., Jahn, D., & Hartig, E. (2006). The Fnr regulon of Bacillus subtilis. Journal of Bacteriology, 188(3), 1103–1112. https://doi.org/10.1128/JB.188.3.1103-1112.2006.
Wei, X., Ji, Z., & Chen, S. (2010). Isolation of halotolerant Bacillus licheniformis WX-02 and regulatory effects of sodium chloride on yield and molecular sizes of poly-gamma-glutamic acid. Applied Biochemistry and Biotechnology, 160(5), 1332–1340. https://doi.org/10.1007/s12010-009-8681-1.
Yangtse, W., Zhou, Y., Lei, Y., Qiu, Y., Wei, X., Ji, Z., Qi, G., Yong, Y., Chen, L., & Chen, S. (2012). Genome sequence of Bacillus licheniformis WX-02. Journal of Bacteriology, 194(13), 3561–3562. https://doi.org/10.1128/JB.00572-12.
Jiang, F., Qi, G., Ji, Z., Zhang, S., Liu, J., Ma, X., & Chen, S. (2011). Expression of glr gene encoding glutamate racemase in Bacillus licheniformis WX-02 and its regulatory effects on synthesis of poly-gamma-glutamic acid. Biotechnology Letters, 33(9), 1837–1840. https://doi.org/10.1007/s10529-011-0631-7.
Tian, G., Fu, J., Wei, X., Ji, Z., Qi, G., & Chen, S. (2013). Enhanced expression of pgdS, gene for high production of poly-γ-glutamic aicd with lower molecular weight in Bacillus licheniformis WX-02. Journal of Chemical Technology and Biotechnology, 89, 1825–1832.
Cai, D., He, P., Lu, X., Zhu, C., Zhu, J., Zhan, Y., Wang, Q., Wen, Z., & Chen, S. (2017). A novel approach to improve poly-γ-glutamic acid production by NADPH regeneration in Bacillus licheniformis WX-02. Scientific Reports, 7, 43404. https://doi.org/10.1038/srep43404.
Wang, J., Yuan, H., Wei, X., Chen, J., & Chen, S. (2015). Enhancement of poly-γ-glutamic acid production by alkaline pH stress treatment in Bacillus licheniformis WX-02. Journal of Chemical Technology and Biotechnology, 121, 1444–1447.
Wei, X., Tian, G., Ji, Z., & Chen, S. (2015). A new strategy for enhancement of poly-γ-glutamic acid production by multiple physicochemical stresses in Bacillus licheniformis WX-02. Journal of Chemical Technology and Biotechnology, 90(4), 709–713. https://doi.org/10.1002/jctb.4362.
Qiu, Y., Zhang, J., Li, L., Wen, Z., Nomura, C. T., Wu, S., & Chen, S. (2016). Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnology for Biofuels, 9(1), 117. https://doi.org/10.1186/s13068-016-0522-1.
Wei, X., Zhou, Y., Chen, J., Cai, D., Wang, D., Qi, G., & Chen, S. (2015). Efficient expression of nattokinase in Bacillus licheniformis: host strain construction and signal peptide optimization. Journal of Industrial Microbiology & Biotechnology, 42(2), 287–295. https://doi.org/10.1007/s10295-014-1559-4.
Cai, D., Wei, X., Qiu, Y., Chen, Y., Chen, J., Wen, Z., & Chen, S. (2016). High-level expression of nattokinase in Bacillus licheniformis by manipulating signal peptide and signal peptidase. Journal of Applied Microbiology, 121(3), 704–712. https://doi.org/10.1111/jam.13175.
Tian, G., Wang, Q., Wei, X., Ma, X., & Chen, S. (2017). Glutamate dehydrogenase (RocG) in Bacillus licheniformis WX-02: enzymatic properties and specific functions in glutamic acid synthesis for poly-γ-glutamic acid production. Enzyme and Microbial Technology, 99, 9–15. https://doi.org/10.1016/j.enzmictec.2017.01.002.
Cataldo, D., Maroon, M., Schrader, L., & Youngs, V. (1975). Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid 1. Commun Soil Sci Plant, 6(1), 71–80. https://doi.org/10.1080/00103627509366547.
Nicholas, D., & Nason, A. (1957). Determination of nitrate and nitrite. Method Enzym, 3, 983–984.
Cai, D., Wang, H., He, P., Zhu, C., Wang, Q., Wei, X., Nomura, C. T., & Chen, S. (2017). A novel strategy to improve protein secretion via overexpression of the SppA signal peptide peptidase in Bacillus licheniformis. Microbial Cell Factories, 16(1), 70. https://doi.org/10.1186/s12934-017-0688-7.
Ogunleye, A., Bhat, A., Irorere, V. U., Hill, D., Williams, C., & Radecka, I. (2015). Poly-gamma-glutamic acid: production, properties and applications. Microbiology, 161(1), 1–17. https://doi.org/10.1099/mic.0.081448-0.
Man, Z., Rao, Z., Xu, M., Guo, J., Yang, T., Zhang, X., & Xu, Z. (2016). Improvement of the intracellular environment for enhancing l-arginine production of Corynebacterium glutamicum by inactivation of H2O2-forming flavin reductases and optimization of ATP supply. Metabolic Engineering, 38, 310–321. https://doi.org/10.1016/j.ymben.2016.07.009.
Bajaj, I., & Singhal, R. (2010). Effect of aeration and agitation on synthesis of poly (γ-glutamic acid) in batch cultures of Bacillus licheniformis NCIM 2324. Biotechnology and Bioprocess Engineering, 15(4), 635–640. https://doi.org/10.1007/s12257-009-0059-2.
Moreno-Vivian, C., Cabello, P., Martinez-Luque, M., Blasco, R., & Castillo, F. (1999). Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. Journal of Bacteriology, 181(21), 6573–6584.
Liu, Y., Zhu, Y., Ma, W., Shin, H. D., Li, J., Liu, L., Du, G., & Chen, J. (2014). Spatial modulation of key pathway enzymes by DNA-guided scaffold system and respiration chain engineering for improved N-acetylglucosamine production by Bacillus subtilis. Metabolic Engineering, 24, 61–69. https://doi.org/10.1016/j.ymben.2014.04.004.
Zamboni, N., Mouncey, N., Hohmann, H. P., & Sauer, U. (2003). Reducing maintenance metabolism by metabolic engineering of respiration improves riboflavin production by Bacillus subtilis. Metabolic Engineering, 5(1), 49–55. https://doi.org/10.1016/S1096-7176(03)00007-7.
Messaoudi, K., Clavel, T., Schmitt, P., & Duport, C. (2010). Fnr mediates carbohydrate-dependent regulation of catabolic and enterotoxin genes in Bacillus cereus F4430/73. Research in Microbiology, 161(1), 30–39. https://doi.org/10.1016/j.resmic.2009.11.003.
Phanaksri, T., Luxananil, P., Panyim, S., & Tirasophon, W. (2015). Synergism of regulatory elements in sigma(B)- and sigma(A)-dependent promoters enhances recombinant protein expression in Bacillus subtilis. Journal of Bioscience and Bioengineering, 120(4), 470–475. https://doi.org/10.1016/j.jbiosc.2015.02.008.
Celinska, E., & Grajek, W. (2009). Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnology Advances, 27(6), 715–725. https://doi.org/10.1016/j.biotechadv.2009.05.002.
Funding
This work was supported by the National Program on Key Basic Research Project (973 Program, No. 2015CB150505) and the Science and Technology Program of Wuhan (20160201010086).
Author information
Authors and Affiliations
Contributions
D Cai and S Chen designed the study. D Cai carried out the molecular biology studies and construction of engineering strains. D Cai, S Hu, Y Chen and L Liu carried out the fermentation studies. D Cai, S Yang, X, Ma and S Chen analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
All the primers sequences for RT-qPCR (Table S1) were supplied as the Supplementary Materials. This information is available free of charge via the Internet http://www.springer.com/chemistry/biotechnology/journal/12010.
ESM 1
(DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Cai, D., Hu, S., Chen, Y. et al. Enhanced Production of Poly-γ-glutamic acid by Overexpression of the Global Anaerobic Regulator Fnr in Bacillus licheniformis WX-02. Appl Biochem Biotechnol 185, 958–970 (2018). https://doi.org/10.1007/s12010-018-2693-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-018-2693-7