Abstract
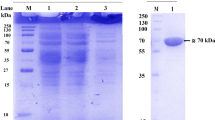
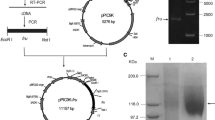
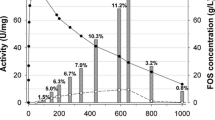
Although fructosyltransferases from Aspergillus aculeatus have received a considerable interest for the prebiotics industry, their amino acid sequences and structural features remain unknown. This study sequenced and characterized a fructosyltransferase from A. aculeatus (AcFT) isolated by heat treatment of Pectinex Ultra SP-L. The AcFT enzyme showed two isoforms, low-glycosylated AcFT1 and high-glycosylated AcFT2 forms, with similar optimum activity at 60 °C. The purified heat-resistant AcFT1 and AcFT2 isoforms produced identical patterns of fructooligosaccharides (FOS; kestose, nystose and fructosylnystose) with a notable transfructosylation capability (~90 % transferase/hydrolase ratio). In contrast, the pI and optimum pH values exhibited discrete differences, attributable to their glycosylation pattern. Partial protein sequencing showed that AcFT enzyme corresponds to Aspac1_37092, a putative 654-residue fructosyltransferase encoded in the genome of A. aculeatus ATCC16872. A homology model of AcFT also revealed the typical fold common to members of the glycoside hydrolase family 32 (GH32), with an N-terminal five-blade β-propeller domain enclosing catalytic residues D60, D191, and E292, linked to a C-terminal β-sandwich domain. To our knowledge, this is the first report describing the amino acid sequence and structural features of a heat-resistant FOS-forming enzyme from A. aculeatus, providing insights into its potential applications in the prebiotics industry.






Similar content being viewed by others
References
Renuka, B., Kulkarni, S. G., Vijayanand, P., & Prapulla, S. G. (2009). Fructooligosaccharide fortification of selected fruit juice beverages: effect on the quality characteristics. LWT - Food Science and Technology, 42(5), 1031–1033.
Flores-Maltos, D. a., Mussatto, S. I., Contreras-Esquivel, J. C., Rodríguez-Herrera, R., Teixeira, J. a., & Aguilar, C. N. (2015). Biotechnological production and application of fructooligosaccharides. Critical Reviews in Biotechnology, 8, 1–9.
Roberfroid, M. B. (2007). Inulin-type fructans: functional food ingredients. The Journal of Nutrition, 137, 2493–2502.
Mussatto, S. I., & Mancilha, I. M. (2007). Non-digestible oligosaccharides: a review. Carbohydrate Polymers, 68(3), 587–597.
Crittenden, R. G., & Playne, M. J. (1996). Production, properties and applications of food-grade oligosaccharides. Trends in Food Science and Technology, 7(11), 353–361.
Antosova, M., & Polakovic, M. (2001). Fructosyltransferases: the enzymes catalyzing production. Chemical Papers, 55(6), 350–358.
Hirayama, M., Sumi, N., & Hidaka, H. (1989). Purification and properties of a fructoooligosaccharide-producing beta-fructofuranosidase from Aspergillus niger ATCC 20611. Agricultural and Biological Chemistry, 53(3), 667–673.
Park, J. P., Oh, T. K., & Yun, J. W. (2001). Purification and characterization of a novel transfructosylating enzyme from Bacillus macerans EG-6. Process Biochemistry, 37(5), 471–476.
Díez-Municio, M., de las Rivas, B., Jimeno, M. L., Muñoz, R., Moreno, F. J., & Herrero, M. (2013). Enzymatic synthesis and characterization of fructooligosaccharides and novel maltosylfructosides by inulosucrase from Lactobacillus gasseri DSM 20604. Applied and Environmental Microbiology, 79(13), 4129–4140.
Ganaie, M. A., Gupta, U. S., & Kango, N. (2013). Screening of biocatalysts for transformation of sucrose to fructooligosaccharides. Journal of Molecular Catalysis B: Enzymatic, 97, 12–17.
Guio, F., Rugeles, L. D., Rojas, S. E., Palomino, M. P., Camargo, M. C., & Sánchez, O. F. (2012). Kinetic modeling of fructooligosaccharide production using Aspergillus oryzae N74. Applied Biochemistry and Biotechnology, 167(1), 142–163.
Lammens, W., Le Roy, K., Schroeven, L., Van Laere, A., Rabijns, A., & Van Den Ende, W. (2009). Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. Journal of Experimental Botany, 60(3), 727–740.
Chuankhayan, P., Hsieh, C. Y., Huang, Y. C., Hsieh, Y. Y., Guan, H. H., Hsieh, Y. C., & Chen, C. J. (2010). Crystal structures of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete subsites in the active site for catalysis. Journal of Biological Chemistry, 285(30), 23251–23264.
Xu, L., Wang, D., Lu, L., Jin, L., Liu, J., Song, D., & Xiao, M. (2014). Purification, cloning, characterization, and N-glycosylation analysis of a novel β-fructosidase from Aspergillus oryzae FS4 synthesizing levan- and neolevan-type fructooligosaccharides. PLoS ONE, 9(12), e114793.
Haraguchi, M., Yamashiro, S., Furukawa, K., Takamiya, K., Shiku, H., & Furukawa, K. (1995). The effects of the site-directed removal of N-glycosylation sites from beta-1,4-N-acetylgalactosaminyltransferase on its function. The Biochemical Journal, 312(Pt1 (1)), 273–280.
Clark, S. E., Muslin, E. H., & Henson, C. a. (2004). Effect of adding and removing N-glycosylation recognition sites on the thermostability of barley β-glucosidase. Protein Engineering Design and Selection, 17(3), 245–249.
Maiorano, A. E., Piccoli, R. M., Da Silva, E. S., & De Andrade Rodrigues, M. F. (2008). Microbial production of fructosyltransferases for synthesis of pre-biotics. Biotechnology Letters, 30(11), 1867–1877.
Muñoz-Gutiérrez, I., Rodríguez-Alegría, M. E., & López Munguía, A. (2009). Kinetic behaviour and specificity of β-fructosidases in the hydrolysis of plant and microbial fructans. Process Biochemistry, 44(8), 891–898.
Lorenzoni, A. S. G., Aydos, L. F., Klein, M. P., Rodrigues, R. C., & Hertz, P. F. (2014). Fructooligosaccharides synthesis by highly stable immobilized β-fructofuranosidase from Aspergillus aculeatus. Carbohydrate Polymers, 103(1), 193–197.
Ghazi, I., Gómez De Segura, A., Fernández-Arrojo, L., Alcalde, M., Yates, M., Rojas-Cervantes, M. L., & Ballesteros, A. (2005). Immobilisation of fructosyltransferase from Aspergillus aculeatus on epoxy-activated Sepabeads EC for the synthesis of fructo-oligosaccharides. Journal of Molecular Catalysis B: Enzymatic, 35(1–3), 19–27.
Adisa, V. a., & Fajola, A. O. (1983). Cellulolytic enzymes associated with the fruit rots of Citrus sinensis caused by Aspergillus aculeatus and Botryodiplodia theobromae. Mycopathologia, 82(1), 23–27.
De Vries, R. P., & Visser, J. (2001). Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiology and Molecular Biology Reviews, 65(4), 497–522.
Vega-Paulino, R. J., & Zúniga-Hansen, M. E. (2012). Potential application of commercial enzyme preparations for industrial production of short-chain fructooligosaccharides. Journal of Molecular Catalysis B: Enzymatic, 76, 44–51.
Ghazi, I., Fernandez-Arrojo, L., Garcia-Arellano, H., Ferrer, M., Ballesteros, A., & Plou, F. J. (2007). Purification and kinetic characterization of a fructosyltransferase from Aspergillus aculeatus. Journal of Biotechnology, 128(1), 204–211.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680–685.
Virgen-Ortíz, J. J., Ibarra-Junquera, V., Osuna-Castro, J. a., Escalante-Minakata, P., Mancilla-Margalli, N. a., & Ornelas-Paz, J. D. J. (2012). Method to concentrate protein solutions based on dialysis-freezing- centrifugation: enzyme applications. Analytical Biochemistry, 426(1), 4–12.
Bradford, M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Miller, G. L. (1959). Use of dinitrosaiicyiic acid reagent for determination of reducing sugar. Analytical Chemistry, 31(3), 426–428.
Nordberg, H., Cantor, M., Dusheyko, S., Hua, S., Poliakov, A., Shabalov, I., & Dubchak, I. (2014). The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Research, 42(D1), 26–31.
Grigoriev, I. V., Nikitin, R., Haridas, S., Kuo, A., Ohm, R., Otillar, R., & Shabalov, I. (2014). MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research, 42(D1), 699–704.
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., & Punta, M. (2014). Pfam: the protein families database. Nucleic Acids Research, 42(D1), 222–230.
Larkin, M. a., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. a., McWilliam, H., & Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21), 2947–2948.
Chuang, G. Y., Boyington, J. C., Gordon Joyce, M., Zhu, J., Nabel, G. J., Kwong, P. D., & Georgiev, I. (2012). Computational prediction of N-linked glycosylation incorporating structural properties and patterns. Bioinformatics, 28(17), 2249–2255.
Nielsen, M., Lundegaard, C., Lund, O., & Petersen, T. N. (2010). CPHmodels-3.0-remote homology modeling using structure-guided sequence profiles. Nucleic Acids Research, 38(SUPPL. 2), 576–581.
Krieger, E., Joo, K., Lee, J., Lee, J., Raman, S., Thompson, J., & Karplus, K. (2009). Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins: Structure, Function, and Bioinformatics, 77(SUPPL. 9), 114–122.
Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. a., Immormino, R. M., Kapral, G. J., & Richardson, D. C. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography, 66(1), 12–21.
Emsley, P., Lohkamp, B., Scott, W. G., & Cowtan, K. (2010). Features and development of Coot. Acta Crystallographica Section D: Biological Crystallography, 66(4), 486–501.
DeBeer, T. a. P., Berka, K., Thornton, J. M., & Laskowski, R. A. (2014). PDBsum additions. Nucleic Acids Research, 42(D1), 292–296. doi:10.1093/nar/gkt940.
McNicholas, S., Potterton, E., Wilson, K. S., & Noble, M. E. M. (2011). Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallographica Section D: Biological Crystallography, 67(4), 386–394.
Sun, F., Zhang, X. Z., Myung, S., & Zhang, Y. H. P. (2012). Thermophilic Thermotoga maritima ribose-5-phosphate isomerase RpiB: optimized heat treatment purification and basic characterization. Protein Expression and Purification, 82(2), 302–307.
Patchett, M. L., Neal, T. L., Schofield, L. R., Strange, R. C., Daniel, R. M., & Morgan, H. W. (1989). Heat treatment purification of thermostable cellulase and hemicellulase enzymes expressed in E. coli. Enzyme and Microbial Technology, 11(2), 113–115.
Gumerov, V. M., Rakitin, A. L., Mardanov, A. V., & Ravin, N. V. (2015). A novel highly thermostable multifunctional beta-glycosidase from Crenarchaeon acidilobus saccharovorans. Archaea, 2015, 978632.
Nguyen, Q. D., Rezessy-Szabó, J. M., Bhat, M. K., & Hoschke, Á. (2005). Purification and some properties of β-fructofuranosidase from Aspergillus niger IMI303386. Process Biochemistry, 40(7), 2461–2466.
Hayashi, S., Matsuzaki, K., Takasaki, Y., Ueno, H., & Imada, K. (1992). Purification and properties of b-fructofuranosidase from Aspergillus japonicus. Microbial Biotechnology, 8, 276–279.
Yanai, K., Nakane, A., Kawate, A., & Hirayama, M. (2001). Molecular cloning and characterization of the fructooligosaccharide-producing beta-fructofuranosidase gene from Aspergillus niger ATCC 20611. Bioscience Biotechnology and Biochemistry, 65, 766–773.
Bruins, M. E., Strubel, M., Van Lieshout, J. F. T., Janssen, A. E. M., & Boom, R. M. (2003). Oligosaccharide synthesis by the hyperthermostable beta-glucosidase from Pyrococcus furiosus: kinetics and modelling. Enzyme and Microbial Technology, 33(1), 3–11.
Zambelli, P., Fernandez-Arrojo, L., Romano, D., Santos-Moriano, P., Gimeno-Perez, M., Poveda, A., & Plou, F. J. (2014). Production of fructooligosaccharides by mycelium-bound transfructosylation activity present in Cladosporium cladosporioides and Penicilium sizovae. Process Biochemistry, 49(12), 2174–2180.
Holm, L., & Rosenström, P. (2010). Dali server: conservation mapping in 3D. Nucleic Acids Research, 38(SUPPL. 2), 545–549.
Hothorn, M., Van den Ende, W., Lammens, W., Rybin, V., & Scheffzek, K. (2010). Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proceedings of the National Academy of Sciences of the United States of America, 107(40), 17427–17432.
Bujacz, A., Jedrzejczak-Krzepkowska, M., Bielecki, S., Redzynia, I., & Bujacz, G. (2011). Crystal structures of the apo form of β-fructofuranosidase from Bifidobacterium longum and its complex with fructose. FEBS Journal, 278(10), 1728–1744.
Pouyez, J., Mayard, A., Vandamme, A. M., Roussel, G., Perpète, E. a., Wouters, J., & Michaux, C. (2012). First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: discovery of an extra-pocket in the catalytic domain responsible for its endo-activity. Biochimie, 94(11), 2423–2430.
Deshpande, N., Wilkins, M. R., Packer, N., & Nevalainen, H. (2008). Protein glycosylation pathways in filamentous fungi. Glycobiology, 18(8), 626–637.
Shental-Bechor, D., & Levy, Y. (2008). Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proceedings of the National Academy of Sciences of the United States of America, 105(24), 8256–8261.
Acknowledgments
This work was supported by the CONACyT, México (No. 169048) and PROMEP. José Juan Virgen-Ortíz would also like to express his gratitude to CONACyT Mexico for his PhD fellowship No. 307333.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Virgen-Ortíz, J.J., Ibarra-Junquera, V., Escalante-Minakata, P. et al. Identification and Functional Characterization of a Fructooligosaccharides-Forming Enzyme from Aspergillus aculeatus . Appl Biochem Biotechnol 179, 497–513 (2016). https://doi.org/10.1007/s12010-016-2009-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2009-8