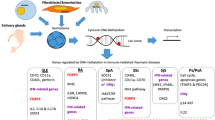
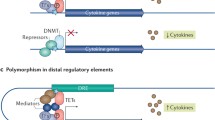
Abstract
Purpose of Review
Epigenetics is the study of inherited phenotype changes that do not involve in alteration of DNA sequence. Epigenetic regulation can be examined under three main headings and study methodologies for all three will be discussed: DNA methylation is the addition of a methyl (CH3) group to DNA, histone modifications is a covalent post-translational modification of histones and non-coding RNAs are a group of functional RNA molecules that is copied from DNA but not converted into proteins. Epigenetic changes are being increasingly studied in the pathogenesis of most diseases including autoimmune and autoinflammatory diseases to shed light on the different phenotypes and disease courses. We have aimed to review the basic concepts in epigenetic studies and summarize the data for epigenetics in autoimmune and autoinflammatory rheumatic diseases.
Recent Findings
Recent studies have assessed DNA hypomethylation in interferon-regulated genes in autoimmune diseases and in inflammatory pathways in systemic autoinflammatory diseases (SAIDs). Abnormal histone acetylation and methylation have been shown to be important in autoimmune diseases which was proven via effective targeted treatment trials against these pathways in mice models. miRNAs have an important role in the pathogenesis, and also, they can be used as diagnostic biomarkers in SAIDs (i.e., FMF, Behcet’s disease) together with autoimmune diseases. Although the number of studies has increased over the years in parallel with the increase of interest in this field, we await further studies to improve the understanding and management of pediatric rheumatic diseases.
Summary
Epigenetic studies in pediatric rheumatic diseases have enabled us to gain new information about disease pathogenesis, clinical heterogeneity, and prognosis. Further studies will help us define new diagnostic, prognostic, and therapeutic goals for rheumatic diseases.


Similar content being viewed by others
Data Availability
Figures were created with BioRender.com.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res - Fundam Mol Mech Mutagen. 1993;285:61–7. https://doi.org/10.1016/0027-5107(93)90052-H.
Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science (80-). 2010;328:916–9. https://doi.org/10.1126/science.1186366.
Zemach A, Zilberman D. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol. 2010;20. https://doi.org/10.1016/j.cub.2010.07.007.
Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6. https://doi.org/10.1101/cshperspect.a019133.
Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: Evidence for X inactivation by DNA methylation. Science (80-). 1981;211:393–6. https://doi.org/10.1126/science.6164095.
Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev. Mol Cell Biol. 2019;20:590–607. https://doi.org/10.1038/s41580-019-0159-6.
Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev. Genet. 2008;9:465–76. https://doi.org/10.1038/nrg2341.
Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, et al. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012;40:4334–46. https://doi.org/10.1093/nar/gks031.
Lim DHK, Maher ER. DNA methylation: a form of epigenetic control of gene expression. Obstet Gynaecol. 2010;12:37–42. https://doi.org/10.1576/toag.12.1.037.27556.
Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. https://doi.org/10.1101/gad.2037511.
Lyko F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat Rev. Genet. 2018;19:81–92. https://doi.org/10.1038/nrg.2017.80.
Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. https://doi.org/10.1016/S0092-8674(00)81656-6.
Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–50. https://doi.org/10.1101/gad.276568.115.
Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–72. https://doi.org/10.1016/j.cell.2011.08.042.
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science (80-). 2011;333:1303–7. https://doi.org/10.1126/science.1210944.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science (80-). 2011;333:1300–3. https://doi.org/10.1126/science.1210597.
• Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–9. https://doi.org/10.1038/nature12750.b. The authors describe perfectly the DNA demethylation enzymes.
Kurdyukov S, Bullock M. DNA methylation analysis: Choosing the right method. Biology (Basel). 2016;5. https://doi.org/10.3390/biology5010003.
Kuo KC, McCune RA, Gehrke CW, Midgett R, Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980;8:4763–76. https://doi.org/10.1093/nar/8.20.4763.
Liu Z, Wu J, Xie Z, Liu S, Fan-Havard P, Huang THM, et al. Quantification of regional DNA methylation by liquid chromatography/tandem mass spectrometry. Anal Biochem. 2009;391:106–13. https://doi.org/10.1016/j.ab.2009.05.012.
Kremer D, Metzger S, Kolb-Bachofen V, Kremer D. Quantitative measurement of genome-wide DNA methylation by a reliable and cost-efficient enzyme-linked immunosorbent assay technique. Anal Biochem. 2012;422:74–8. https://doi.org/10.1016/j.ab.2011.11.033.
Mohsen K, Johansson S, Ekström TJ, Karimi M. Using LUMA: a luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–8. https://doi.org/10.4161/epi.1.1.2587.
Karimi M, Johansson S, Stach D, Corcoran M, Grandér D, Schalling M, et al. LUMA (LUminometric Methylation Assay)-a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–95. https://doi.org/10.1016/j.yexcr.2006.03.006.
Schumacher A, Kapranov P, Kaminsky Z, Flanagan J, Assadzadeh A, Yau P, et al. Microarray-based DNA methylation profiling: technology and applications. Nucleic Acids Res. 2006;34:528–42. https://doi.org/10.1093/nar/gkj461.
Li Y, Tollefsbol TO. DNA methylation detection: bisulfite genomic sequencing analysis. Methods Mol Biol. 2011;791:11–21. https://doi.org/10.1007/978-1-61779-316-5_2.
Angelini F, Pagano F, Bordin A, Milan M, Chimenti I, Peruzzi M, et al. The impact of environmental factors in influencing epigenetics related to oxidative states in the cardiovascular System. Oxid Med Cell Longev. 2017;2017. https://doi.org/10.1155/2017/2712751.
Akbaba T, Balcı-Peynircioğlu B. Potential impacts of gut microbiota on immune system related diseases: current studies and future challenges. Acta Medica Cordoba. 2018;49:31–7.
Mizugaki M, Yamaguchi T, Ishiwata S, Shindo H, Hishinuma T, Nozaki S, et al. Alteration of DNA methylation levels in MRL lupus mice. Clin Exp Immunol. 1997;110:265–9. https://doi.org/10.1111/j.1365-2249.1997.tb08326.x.
Zhou Y, Lu Q. DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmun Rev. 2008;7:376–83. https://doi.org/10.1016/j.autrev.2008.03.003.
Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–73. https://doi.org/10.1002/art.1780331109.
Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–75. https://doi.org/10.1016/s1074-7613(00)80642-1.
Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–73. https://doi.org/10.4049/jimmunol.182.1.259.
Ngalamika O, Liang G, Zhao M, Yu X, Yang Y, Yin H, et al. Peripheral whole blood FOXP3 TSDR methylation: a potential marker in severity assessment of autoimmune diseases and chronic infections. Immunol Invest. 2015;44:126–36. https://doi.org/10.3109/08820139.2014.938165.
Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–9. https://doi.org/10.1101/gr.100289.109.
Zhao M, Liu S, Luo S, Wu H, Tang M, Cheng W, et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J Autoimmun. 2014;54:127–36. https://doi.org/10.1016/j.jaut.2014.07.002.
Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84. https://doi.org/10.1016/j.jaut.2013.04.003.
Zhao M, Zhou Y, Zhu B, Wan M, Jiang T, Tan Q, et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann Rheum Dis. 2016;75:1998–2006. https://doi.org/10.1136/annrheumdis-2015-208410.
Imgenberg-Kreuz J, Carlsson Almlof J, Leonard D, Alexsson A, Nordmark G, Eloranta ML, et al. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann Rheum Dis. 2018;77:736–43. https://doi.org/10.1136/annrheumdis-2017-212379.
Coit P, Renauer P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, et al. Renal involvement in lupus is characterized by unique DNA methylation changes in naive CD4+ T cells. J Autoimmun. 2015;61:29–35. https://doi.org/10.1016/j.jaut.2015.05.003.
Renauer P, Coit P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, et al. DNA methylation patterns in naive CD4+ T cells identify epigenetic susceptibility loci for malar rash and discoid rash in systemic lupus erythematosus. Lupus Sci Med. 2015;2:e000101. https://doi.org/10.1136/lupus-2015-000101.
Li Y, Zhao M, Yin H, Gao F, Wu X, Luo Y, et al. Overexpression of the growth arrest and DNA damage-induced 45alpha gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. 2010;62:1438–47. https://doi.org/10.1002/art.27363.
Li Y, Huang C, Zhao M, Liang G, Xiao R, Yung S, et al. A possible role of HMGB1 in DNA demethylation in CD4+ T cells from patients with systemic lupus erythematosus. Clin Dev Immunol. 2013;2013:206298. https://doi.org/10.1155/2013/206298.
Zhao M, Sun Y, Gao F, Wu X, Tang J, Yin H, et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J Autoimmun. 2010;35:58–69. https://doi.org/10.1016/j.jaut.2010.02.002.
Zhao M, Wang J, Liao W, Li D, Li M, Wu H, et al. Increased 5-hydroxymethylcytosine in CD4(+) T cells in systemic lupus erythematosus. J Autoimmun. 2016;69:64–73. https://doi.org/10.1016/j.jaut.2016.03.001.
Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–26. https://doi.org/10.1016/j.immuni.2015.03.005.
Fali T, Le Dantec C, Thabet Y, Jousse S, Hanrotel C, Youinou P, et al. DNA methylation modulates HRES1/p28 expression in B cells from patients with Lupus. Autoimmunity. 2014;47:265–71. https://doi.org/10.3109/08916934.2013.826207.
Nakkuntod J, Avihingsanon Y, Mutirangura A, Hirankarn N. Hypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patients. Clin Chim Acta. 2011;412:1457–61. https://doi.org/10.1016/j.cca.2011.04.002.
Scharer CD, Blalock EL, Mi T, Barwick BG, Jenks SA, Deguchi T, et al. Epigenetic programming underpins B cell dysfunction in human SLE. Nat Immunol. 2019;20:1071–82. https://doi.org/10.1038/s41590-019-0419-9.
Kirectepe AK, Kasapcopur O, Arisoy N, Celikyapi Erdem G, Hatemi G, Ozdogan H, et al. Analysis of MEFV exon methylation and expression patterns in familial Mediterranean fever. BMC Med Genet. 2011;12:105. https://doi.org/10.1186/1471-2350-12-105.
Vento-Tormo R, Álvarez-Errico D, Garcia-Gomez A, Hernández-Rodríguez J, Buján S, Basagaña M, et al. DNA demethylation of inflammasome-associated genes is enhanced in patients with cryopyrin-associated periodic syndromes. J Allergy Clin Immunol. 2017;139:202–211.e6. https://doi.org/10.1016/j.jaci.2016.05.016.
Hughes T, Ture-Ozdemir F, Alibaz-Oner F, Coit P, Direskeneli H, Sawalha AH. Epigenome-wide scan identifies a treatment-responsive pattern of altered DNA methylation among cytoskeletal remodeling genes in monocytes and CD4+ T cells from patients with Behcet’s disease. Arthritis Rheumatol. 2014;66:1648–58. https://doi.org/10.1002/art.38409.
Hu N, Qiu X, Luo Y, Yuan J, Li Y, Lei W, et al. Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol. 2008;35:804–10.
Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 2010;11:124–33. https://doi.org/10.1038/gene.2009.66.
Dai Y, Zhang L, Hu C, Zhang Y. Genome-wide analysis of histone H3 lysine 4 trimethylation by ChIP-chip in peripheral blood mononuclear cells of systemic lupus erythematosus patients. Clin Exp Rheumatol. 2010;28:158–68.
Zhao H, Wang L, Luo H, Li QZ, Zuo X. TNFAIP3 downregulation mediated by histone modification contributes to T cell dysfunction in systemic lupus erythematosus. Rheumatol. 2017;56:835–43. https://doi.org/10.1093/rheumatology/kew508.
Sullivan KE, Suriano A, Dietzmann K, Lin J, Goldman D, Petri MA. The TNFalpha locus is altered in monocytes from patients with systemic lupus erythematosus. Clin Immunol. 2007;123:74–81. https://doi.org/10.1016/j.clim.2006.12.008.
Apostolidis SA, Rauen T, Hedrich CM, Tsokos GC, Crispin JC. Protein phosphatase 2A enables expression of interleukin 17 (IL-17) through chromatin remodeling. J Biol Chem. 2013;288:26775–84. https://doi.org/10.1074/jbc.M113.483743.
Hedrich CM, Rauen T, Apostolidis SA, Grammatikos AP, Rodriguez Rodriguez N, Ioannidis C, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc Natl Acad Sci U S A. 2014;111:13457–62. https://doi.org/10.1073/pnas.1408023111.
Wardowska A, Komorniczak M, Bullo-Piontecka B, Debska-Slizien MA, Pikula M. Transcriptomic and epigenetic alterations in dendritic cells correspond with chronic kidney disease in lupus nephritis. Front Immunol. 2019;10:2026. https://doi.org/10.3389/fimmu.2019.02026.
Rother N, Pieterse E, Lubbers J, Hilbrands L, van der Vlag J. Acetylated histones in apoptotic microparticles drive the formation of neutrophil extracellular traps in active lupus nephritis. Front Immunol. 2017;8:1136. https://doi.org/10.3389/fimmu.2017.01136.
Hofmann SR, Kubasch AS, Ioannidis C, Rösen-Wolff A, Girschick HJ, Morbach H, et al. Altered expression of IL-10 family cytokines in monocytes from CRMO patients result in enhanced IL-1β expression and release. Clin Immunol. 2015;161:300–7. https://doi.org/10.1016/j.clim.2015.09.013.
Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–81. https://doi.org/10.4049/jimmunol.0904060.
Rouas R, Fayyad-Kazan H, El Zein N, Lewalle P, Rothe F, Simion A, et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol. 2009;39:1608–18. https://doi.org/10.1002/eji.200838509.
Stagakis E, Bertsias G, Verginis P, Nakou M, Hatziapostolou M, Kritikos H, et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis. 2011;70:1496–506. https://doi.org/10.1136/ard.2010.139857.
Liao Z, Ye Z, Xue Z, Wu L, Ouyang Y, Yao C, et al. Identification of renal long non-coding RNA RP11-2B6.2 as a positive regulator of type I interferon signaling pathway in lupus nephritis. Front Immunol. 2019;10:975. https://doi.org/10.3389/fimmu.2019.00975.
Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B, et al. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res Ther. 2015;17:131. https://doi.org/10.1186/s13075-015-0632-3.
Latsoudis H, Mashreghi MF, Grün JR, Chang HD, Stuhlmüller B, Repa A, et al. Differential expression of mir-4520a associated with pyrin mutations in familial Mediterranean fever (FMF). J Cell Physiol. 2017;232:1326–36. https://doi.org/10.1002/jcp.25602.
Koga T, Migita K, Sato T, Sato S, Umeda M, Nonaka F, et al. MicroRNA-204-3p inhibits lipopolysaccharide-induced cytokines in familial Mediterranean fever via the phosphoinositide 3-kinase gamma pathway. Rheumatol. 2018;57:718–26. https://doi.org/10.1093/rheumatology/kex451.
Akkaya-Ulum YZ, Balci-Peynircioglu B, Karadag O, Eroglu FK, Kalyoncu U, Kiraz S, et al. Alteration of the microRNA expression profile in familial Mediterranean fever patients. Clin Exp Rheumatol. 2017;35(Suppl 1):90–4.
Amarilyo G, Pillar N, Ben-Zvi I, Weissglas-Volkov D, Zalcman J, Harel L, et al. Analysis of microRNAs in familial Mediterranean fever. PLoS One. 2018;13:e0197829. https://doi.org/10.1371/journal.pone.0197829.
Akkaya-Ulum ZY, Tavukcuoglu Z, Batu ED, Akbaba TH, Sonmez HS, Balci-Peynircioglu B. et al. Possible regulatory effects of miRNAs in the pathogenesis of systemic auto inflammatory diseases, from the perspective of familial Mediterranean fever. 10th Congress of International Society of Systemic Auto Inflammatory Diseases (ISSAID). Pediatr Rheumatol. 2019;17:18. https://doi.org/10.1186/s12969-019-0313-x.
Oner T, Yenmis G, Tombulturk K, Cam C, Kucuk OS, Yakicier MC, et al. Association of pre-miRNA-499 rs3746444 and pre-miRNA-146a rs2910164 polymorphisms and susceptibility to Behcet’s disease. Genet Test Mol Biomarkers. 2015;19:424–30. https://doi.org/10.1089/gtmb.2015.0016.
Ibrahim W, Sakr BR, Obaya E, Ghonem H. MicroRNA-146a expression and microRNA-146a rs2910164 polymorphism in Behcet’s disease patients. Clin Rheumatol. 2019;38:397–402. https://doi.org/10.1007/s10067-018-4191-2.
Zhou Q, Hou S, Liang L, Li X, Tan X, Wei L, et al. MicroRNA-146a and Ets-1 gene polymorphisms in ocular Behcet’s disease and Vogt-Koyanagi-Harada syndrome. Ann Rheum Dis. 2014;73:170–6. https://doi.org/10.1136/annrheumdis-2012-201627.
Yu H, Liu Y, Bai L, Kijlstra A, Yang P. Predisposition to Behcet’s disease and VKH syndrome by genetic variants of miR-182. J Mol Med. 2014;92:961–7. https://doi.org/10.1007/s00109-014-1159-9.
Qi J, Hou S, Zhang Q, Liao D, Wei L, Fang J, et al. A functional variant of pre-miRNA-196a2 confers risk for Behcet’s disease but not for Vogt-Koyanagi-Harada syndrome or AAU in ankylosing spondylitis. Hum Genet. 2013;132:1395–404. https://doi.org/10.1007/s00439-013-1346-8.
Lee KK, Workman JL. Histone acetyltransferase complexes: One size does not fit all. Nat Rev. Mol Cell Biol. 2007;8:284–95. https://doi.org/10.1038/nrm2145.
Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. https://doi.org/10.1038/cr.2011.22.
Oliver SS, Denu JM. Dynamic interplay between histone H3 modifications and protein interpreters: emerging evidence for a “histone language”. ChemBioChem. 2011;12:299–307. https://doi.org/10.1002/cbic.201000474.
Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–59. https://doi.org/10.1128/mmbr.64.2.435-459.2000.
Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol. 2006;26:6357–71. https://doi.org/10.1128/MCB.00311-06.
Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Respir J. 2005;25:552–63. https://doi.org/10.1183/09031936.05.00117504.
Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6. https://doi.org/10.1101/cshperspect.a018713.
Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev. Genet. 2012;13:343–57. https://doi.org/10.1038/nrg3173.
Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49. https://doi.org/10.1038/emm.2017.11.
Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev. Mol Cell Biol. 2005;6:838–49. https://doi.org/10.1038/nrm1761.
Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol. 2015;6. https://doi.org/10.3389/fimmu.2015.00487.
Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. https://doi.org/10.1016/j.molcel.2012.11.006.
Rossetto D, Avvakumov N, Côté J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7:1098–108. https://doi.org/10.4161/epi.21975.
Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2003;100:13225–30. https://doi.org/10.1073/pnas.1735528100.
Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells. Eur J Biochem. 2001;268:5424–9. https://doi.org/10.1046/j.0014-2956.2001.02481.x.
Lara E, Calvanese V, Fernandez AF, Fraga MF. Techniques to study DNA methylation and histone modification. Epigenetic Asp. Chronic Dis., Springer London. 2011:21–39. https://doi.org/10.1007/978-1-84882-644-1_2.
Song N, Liu J, An S, Nishino T, Hishikawa Y, Koji T. Immunohistochemical analysis of histone H3 modifications in germ cells during mouse spermatogenesis. Acta Histochem Cytochem. 2011;44:183–90. https://doi.org/10.1267/ahc.11027.
Collas P. The current state of chromatin immunoprecipitation. Mol Biotechnol. 2010;45:87–100. https://doi.org/10.1007/s12033-009-9239-8.
• Sidoli S, Bhanu NV, Karch KR, Wang X, Garcia BA. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J Vis Exp. 2016;2016. https://doi.org/10.3791/54112. The authors explained the workflow of the MS-based method for histone post-transcriptional modification studies.
Benedetti R, Conte M, Altucci L. Targeting histone deacetylases in diseases: where are we? Antioxidants Redox Signal. 2015;23:99–126. https://doi.org/10.1089/ars.2013.5776.
Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. 2003;111:539–52. https://doi.org/10.1172/JCI16153.
Hu N, Long H, Zhao M, Yin H, Lu Q. Aberrant expression pattern of histone acetylation modifiers and mitigation of lupus by SIRT1-siRNA in MRL/lpr mice. Scand J Rheumatol. 2009;38:464–71. https://doi.org/10.3109/03009740902895750.
Shu J, Li L, Zhou LB, Qian J, Fan ZD, Zhuang LL, et al. IRF5 is elevated in childhood-onset SLE and regulated by histone acetyltransferase and histone deacetylase inhibitors. Oncotarget. 2017;8:47184–94. https://doi.org/10.18632/oncotarget.17586.
Choi EW, Song JW, Ha N, Choi YI, Kim S. CKD-506, a novel HDAC6-selective inhibitor, improves renal outcomes and survival in a mouse model of systemic lupus erythematosus. Sci Rep. 2018;8:17297. https://doi.org/10.1038/s41598-018-35602-1.
Ren J, Catalina MD, Eden K, Liao X, Read KA, Luo X, et al. Selective histone deacetylase 6 inhibition normalizes B cell activation and germinal center formation in a model of systemic lupus erythematosus. Front Immunol. 2019;10:2512. https://doi.org/10.3389/fimmu.2019.02512.
Ren J, Liao X, Vieson MD, Chen M, Scott R, Kazmierczak J, et al. Selective HDAC6 inhibition decreases early stage of lupus nephritis by down-regulating both innate and adaptive immune responses. Clin Exp Immunol. 2018;191:19–31. https://doi.org/10.1111/cei.13046.
Carta S, Tassi S, Semino C, Fossati G, Mascagni P, Dinarello CA, et al. Histone deacetylase inhibitors prevent exocytosis of interleukin-1beta-containing secretory lysosomes: role of microtubules. Blood. 2006;108:1618–26. https://doi.org/10.1182/blood-2006-03-014126.
Leoni F, Fossati G, Lewis EC, Lee JK, Porro G, Pagani P, et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11:1–15. https://doi.org/10.2119/2006-00005.Dinarello.
Bodar EJ, Simon A, van der Meer JW. Effects of the histone deacetylase inhibitor ITF2357 in autoinflammatory syndromes. Mol Med. 2011;17:363–8. https://doi.org/10.2119/molmed.2011.00039.
Palazzo AF, Lee ES. Non-coding RNA: What is functional and what is junk? Front Genet. 2015;5. https://doi.org/10.3389/fgene.2015.00002.
Hombach S, Kretz M. Non-coding RNAs: Classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. https://doi.org/10.1007/978-3-319-42059-2_1.
MacFarlane L-A, Murphy RP. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–61. https://doi.org/10.2174/138920210793175895.
Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–62. https://doi.org/10.1093/nar/gky1141.
Hashimoto Y, Akiyama Y, Yuasa Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS One. 2013;8. https://doi.org/10.1371/journal.pone.0062589.
• Gu W, Xu Y, Xie X, Wang T, Ko JH, Zhou T. The role of RNA structure at 5′ untranslated region in microRNA-mediated gene regulation. RNA. 2014;20:1369–75. https://doi.org/10.1261/rna.044792.114. Authors identified the role of the 5′ untranslated region as a previously under-appreciated epigenetic mechanism in gene regulation.
Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev. RNA. 2012;3:311–30. https://doi.org/10.1002/wrna.121.
Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21:542–51. https://doi.org/10.1038/s41556-019-0311-8.
Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. https://doi.org/10.1101/gr.3715005.
Cabili M, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. https://doi.org/10.1101/gad.17446611.
Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. https://doi.org/10.1016/0092-8674(92)90519-I.
Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. https://doi.org/10.1038/sj.onc.1206928.
Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. https://doi.org/10.1128/mcb.10.1.28.
Forero DA, González-Giraldo Y, Castro-Vega LJ, Barreto GE. qPCR-based methods for expression analysis of miRNAs. Biotechniques. 2019;67:192–9. https://doi.org/10.2144/btn-2019-0065.
Hunt EA, Broyles D, Head T, Deo SK. MicroRNA Detection: Current Technology and Research Strategies. Annu Rev. Anal Chem. 2015;8:217–37. https://doi.org/10.1146/annurev-anchem-071114-040343.
• Biscontin A, Casara S, Cagnin S, Tombolan L, Rosolen A, Lanfranchi G, et al. New miRNA labeling method for bead-based quantification. BMC Mol Biol. 2010;11. https://doi.org/10.1186/1471-2199-11-44. The authors investigated and defined a new and easy method for miRNA studies.
Balci-Peynircioglu B, Akkaya-Ulum YZ, Akbaba TH, Tavukcuoglu Z. Potential of miRNAs to predict and treat inflammation from the perspective of familial Mediterranean fever. Inflamm Res. 2019;68:905–13. https://doi.org/10.1007/s00011-019-01272-6.
Feng Y, Hu X, Zhang Y, Zhang D, Li C, Zhang L. Methods for the study of long noncoding RNA in cancer cell signaling. Methods Mol Biol. 2014;1165:115–43. https://doi.org/10.1007/978-1-4939-0856-1_10.
Cao M, Zhao J, Hu G. Genome-wide methods for investigating long noncoding RNAs. Biomed Pharmacother. 2019;111:395–401. https://doi.org/10.1016/j.biopha.2018.12.078.
Mongelli A, Martelli F, Farsetti A, Gaetano C. The dark that matters: Long noncoding RNAs as master regulators of cellular metabolism in noncommunicable diseases. Front Physiol. 2019;10. https://doi.org/10.3389/fphys.2019.00369.
Tang Y, Zhou T, Yu X, Xue Z, Shen N. The role of long non-coding RNAs in rheumatic diseases. Nat Rev. Rheumatol. 2017;13:657–69. https://doi.org/10.1038/nrrheum.2017.162.
Zheng X, Zhang Y, Yue P, Liu L, Wang C, Zhou K, et al. Diagnostic significance of circulating miRNAs in systemic lupus erythematosus. PLoS One. 2019;14:e0217523. https://doi.org/10.1371/journal.pone.0217523.
Ye H, Wang X, Wang L, Chu X, Hu X, Sun L, et al. Full high-throughput sequencing analysis of differences in expression profiles of long noncoding RNAs and their mechanisms of action in systemic lupus erythematosus. Arthritis Res Ther. 2019;21:70. https://doi.org/10.1186/s13075-019-1853-7.
Wang Y, Chen S, Chen S, Du J, Lin J, Qin H, et al. Long noncoding RNA expression profile and association with SLEDAI score in monocyte-derived dendritic cells from patients with systematic lupus erythematosus. Arthritis Res Ther. 2018;20:138. https://doi.org/10.1186/s13075-018-1640-x.
Akkaya-Ulum ZY, Tavukcuoglu Z, Akbaba TH, Yılmaz E, Balci-Peynircioglu B. miR-197 regulates inflammation in monocytes and synovial fibroblasts by targeting IL1R1.10th Congress of International Society of Systemic Auto Inflammatory Diseases (ISSAID). Pediatr Rheumatol. 2019:17. https://doi.org/10.1186/s12969-019-0313-x.
Hortu HO, Karaca E, Sozeri B, Gulez N, Makay B, Gunduz C, et al. Evaluation of the effects of miRNAs in familial Mediterranean fever. Clin Rheumatol. 2019;38:635–43. https://doi.org/10.1007/s10067-017-3914-0.
Zhou Q, Xiao X, Wang C, Zhang X, Li F, Zhou Y, et al. Decreased microRNA-155 expression in ocular Behcet’s disease but not in Vogt Koyanagi Harada syndrome. Invest Ophthalmol Vis Sci. 2012;53:5665–74. https://doi.org/10.1167/iovs.12-9832.
Ahmadi M, Yousefi M, Abbaspour-Aghdam S, Dolati S, Aghebati-Maleki L, Eghbal-Fard S, et al. Disturbed Th17/Treg balance, cytokines, and miRNAs in peripheral blood of patients with Behcet’s disease. J Cell Physiol. 2019;234:3985–94. https://doi.org/10.1002/jcp.27207.
Na SY, Park MJ, Park S, Lee ES. MicroRNA-155 regulates the Th17 immune response by targeting Ets-1 in Behcet’s disease. Clin Exp Rheumatol. 2016;34:S56–63.
Kolahi S, Farajzadeh MJ, Alipour S, Abhari A, Farhadi J, Bahavarnia N, et al. Determination of mir-155 and mir-146a expression rates and its association with expression level of TNF-alpha and CTLA4 genes in patients with Behcet’s disease. Immunol Lett. 2018;204:55–9. https://doi.org/10.1016/j.imlet.2018.10.012.
Consent for Publication
All authors agree on publishing.
Funding
This study is a part of the ERARE3 project (INSAID, grant 003037603) and funded by the Technical and Scientific Research Council of Turkey (TUBITAK), grant number: 315S096.
Author information
Authors and Affiliations
Contributions
THA wrote the manuscript and made figures. ES wrote the manuscript. BBP and SO reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Pediatric Rheumatology
Rights and permissions
About this article
Cite this article
Akbaba, T.H., Sag, E., Balci-Peynircioglu, B. et al. Epigenetics for Clinicians from the Perspective of Pediatric Rheumatic Diseases. Curr Rheumatol Rep 22, 46 (2020). https://doi.org/10.1007/s11926-020-00912-9
Published:
DOI: https://doi.org/10.1007/s11926-020-00912-9