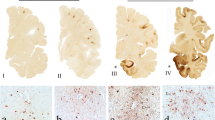
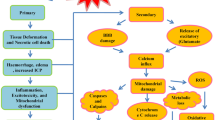
Abstract
Traumatic brain injury (TBI) and traumatic spinal cord injury (SCI), collectively termed neurotrauma, are two parallel neurological conditions that can cause long-lasting neurological impairment and other comorbidities in patients, while at the same time, can create a high burden to society. To date, there are still no FDA-approved therapeutic interventions for either TBI or SCI. Recent advances in proteomic technologies, including tandem mass spectrometry, as well as imaging mass spectrometry, have enabled new approaches to study the differential proteome in TBI and SCI with the use of either animal disease models and/or biosamples from clinical observational studies. Thus, the applications of state-of-the-art proteomic method hold promises in shedding light on identifying clinically useful neurotrauma “biomarkers” and/or in identifying distinct and, otherwise, unobvious systems pathways or “key drivers” that can be further exploited as new therapeutic intervention targets.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Coles JP, Fryer TD, Smielewski P, Chatfield DA, Steiner LA, Johnston AJ, et al. Incidence and mechanisms of cerebral ischemia in early clinical head injury. J Cereb Blood Flow Metab. 2004;24:202–11.
Menon DK, Coles JP, Gupta AK, Fryer TD, Smielewski P, Chatfield DA, et al. Diffusion limited oxygen delivery following head injury. Crit Care Med. 2004;32(6):1384–90.
Lakshmanan R, Loo JA, Drake T, Leblanc J, Ytterberg AJ, Mcarthur DL, et al. Metabolic crisis after traumatic brain injury is associated with a novel microdialysis proteome. Neurocrit Care. 2010;12(3):324–36.
Timofeev I, Carpenter KLH, Nortje J, Al-Rawi PG, O’connell MT, Czosnyka M, et al. Cerebral extracellular chemistry and outcome following traumatic brain injury: a microdialysis study of 223 patients. Brain. 2011;134(2):484–94.
Zhang P, Zhu S, Li Y, Zhao M, Liu M, Gao J et al. Quantitative proteomics analysis to identify diffuse axonal injury biomarkers in rats using iTRAQ coupled LC–MS/MS. J Proteomics. 2016;133:93–9.
Kumar A, Loane DJ. Neuroinflammation after traumatic brain injury: opportunities for therapeutic intervention. Brain Behav Immun. 2012;26(8):1191–201.
Raghupathi R. Cell death mechanisms following traumatic brain injury. Brain Pathol. 2004;14(2):215–22.
• Chauhan NB. Chronic neurodegenerative consequences of traumatic brain injury. Restor Neurol Neurosci. 2014;32:337–65. Chauhan studied the consequences of traumatic brain injuries and its wide-ranging possibilities of affecting people’s lives. He studied common and deadly diseases such as Alzheimer’s and its relationship to TBI and repeated TBI can lead to chronic traumatic encephalopathy (CTE).
Rao VLR, Dhodda VK, Song G, Bowen KK, Dempsey RJ. Traumatic brain injury-induced acute gene expression changes in rat cerebral cortex identified by GeneChip analysis. J Neurosci Res. 2003;71(2):208–19.
Wang KKW. Calpain and caspase: can you tell the difference? Trends Neurosci. 2000;23(1):20–6.
Ansari MA, Roberts KN, Scheff SW. Oxidative stress and modification of synaptic proteins in hippocampus after traumatic brain injury. Free Radic Biol Med. 2008;45(4):443–52.
Smith DH, Uryu K, Saatman KE, Trojanowski JQ, McIntosh TK. Protein accumulation in traumatic brain injury. Neuromolecular Med. 2003;4:59–72.
Zhang X, Chen Y, Ikonomovic MD, Nathaniel PD, Kochanek PM, Marion DW, et al. Increased phosphorylation of protein kinase B and related substrates after traumatic brain injury in humans and rats. J Cereb Blood Flow Metab. 2005;26(7):915–26.
Yang Z, Wang KK. Glial fibrillary acidic protein: from intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 2015;38(6):364–74.
Rosenberg GA. Matrix metalloproteinases in brain injury. J Neurotrauma. 1995;12(5):833–42.
Hook G, Jacobsen JS, Grabstein K, Kindy M, Hook V. Cathepsin B is a new drug target for traumatic brain injury therapeutics: evidence for E64d as a promising lead drug candidate. Front Neurol. 2015;6.
Faul M, Xu L, Wald MM, Coronado VG. Traumatic brain injury in the United States emergency department visits, hospitalizations and deaths 2002–2006. 2010;1–74.
•• Ordikhani F, Sheth S, Zustiak SP. Polymeric particle-mediated molecular therapies to treat spinal cord injury. Int J Pharm. 2016;16(31073):S0378–5173. Ordikhani et al. recognizes the issues that affect those that suffer from SCI. They review the benefits and challenges that polymeric particles can take on as a potential therapeutic help to those needing SCI treatment.
National Center for Injury Prevention and Control. Report to congress on mild traumatic brain injury in the united states: steps to prevent a serious public health problem. 2003;1–45.
Yamasaki K, Setoguchi T, Takenouchi T, Yone K, Komiya S. Stem cell factor prevents neuronal cell apoptosis after acute spinal cord injury. Spine. 2009;34(4):323–7.
Chen M, Xia X, Zhu X, Cao J, Xu D, Ni Y, et al. Expression of SGTA correlates with neuronal apoptosis and reactive gliosis after spinal cord injury. Cell Tissue Res. 2014;358(2):277–88.
Ellis RC, Earnhardt JN, Hayes RL, Wang KK, Anderson DK. Cathepsin B mRNA and protein expression following contusion spinal cord injury in rats. J Neurochem. 2004;88(3):689–97.
Ellis RC, O’steen WA, Hayes RL, Nick HS, Wang KK, Anderson DK. Cellular localization and enzymatic activity of cathepsin B after spinal cord injury in the rat. Exp Neurol. 2005;193(1):19–28.
Zhang H, Chu G, Pan C, Hu J, Guo C, Liu J, et al. A nutrient mixture reduces the expression of matrix metalloproteinases in an animal model of spinal cord injury by modulating matrix metalloproteinase-2 and matrix metalloproteinase-9 promoter activities. Exp Ther Med. 2014;8(6):1835–40.
Goldstein M. Traumatic brain injury: a silent epidemic. Ann Neurol. 1990;27(3):327–7.
Hoffman SW, Harrison C. The interaction between psychological health and traumatic brain injury: a neuroscience perspective. Clin Neuropsychol. 2009;23(8):1400–15.
Papurica M, Rogobete AF, Sandesc D, Dumache R, Cradigati CA, Sarandan M, et al. Advances in biomarkers in critical ill polytrauma patients. Clin Lab. 2016;62(6):977–86.
Stammers A, Liu J, Kwon B. Expression of inflammatory cytokines following acute spinal cord injury in a rodent model. J Neurosci Res. 2012;90(4):782–90.
Kwon BK, Stammers AM, Belanger LM, Bernardo A, Chan D, Bishop CM, et al. Cerebrospinal fluid inflammatory cytokines and biomarkers of injury severity in acute human spinal cord injury. J Neurotrauma. 2010;27(4):669–82.
Kwon BK, Casha S, Hurlbert RJ, Yong VW. Inflammatory and structural biomarkers in acute traumatic spinal cord injury. Clin Chem Lab Med. 2011;49(3):425–33.
Springer JE, Azbill RD, Kennedy SE, George J, Geddes JW. Rapid calpain I activation and cytoskeletal protein degradation following traumatic spinal cord injury: attenuation with riluzole pretreatment. J Neurochem. 1997;69(4):1592–600.
Zhang SX, Underwood M, Landfield A, Huang F-F, Gibson S, Geddes JW. Cytoskeletal disruption following contusion injury to the rat spinal cord. J Neuropathol Exp Neurol. 2000;59(4):287–96.
Hanrieder J, Wetterhall M, Enblad P, Hillered L, Bergquist J. Temporally resolved differential proteomic analysis of human ventricular CSF for monitoring traumatic brain injury biomarker candidates. J Neurosci Methods. 2009;177(2):469–78.
•• Xu B, Tian R, Wang X, Zhan S, Wang R, Guo Y, et al. Protein profile changes in the frontotemporal lobes in human severe traumatic brain injury. Brain Res. 2016;1642:344–52. Xu et al. utilized bioinformatics analysis to relate the effects of severe traumatic brain injury with alterations in biological pathways in the brain. They observed through both mass spectrometry and western blot techniques the increased levels of GFAP, MMP9, transgelin, and vimentin in people suffering from severe TBI (sTBI).
Lööv C, Shevchenko G, Nadadhur AG, Clausen F, Hillered L, Wetterhall M et al. Identification of injury specific proteins in a cell culture model of traumatic brain injury. PLoS One. 2013;8(2).
Ahmed F, Gyorgy A, Kamnaksh A, Ling G, Tong L, Parks S, et al. Time-dependent changes of protein biomarker levels in the cerebrospinal fluid after blast traumatic brain injury. Electrophoresis. 2012;33(24):3705–11.
Kobeissy FH, Sadasivan S, Liu J, Gold MS, Wang KK. Psychiatric research: psychoproteomics, degradomics and systems biology. Expert Rev Proteomics. 2008;5(2):293–314.
Wu P, Zhao Y, Haidacher SJ, Wang E, Parsley MO, Gao J, et al. Detection of structural and metabolic changes in traumatically injured hippocampus by quantitative differential proteomics. J Neurotrauma. 2013;30(9):775–88.
Alder J, Fujioka W, Lifshitz J, Crockett DP, Thakker-Varia S. Lateral fluid percussion: model of traumatic brain injury in mice. J Vis Exp. 2011;(54).
Reed TT, Jr WMP, Turner DM, Markesbery WR, Butterfield DA. Proteomic identification of nitrated brain proteins in early Alzheimer’s disease inferior parietal lobule. J Cell Mol Med. 2009;13(8b):2019–29.
Guingab-Cagmat JD, Newsom K, Vakulenko A, Cagmat EB, Kobeissy FH, Zoltewicz S, et al. In vitro MS-based proteomic analysis and absolute quantification of neuronal-glial injury biomarkers in cell culture system. Electrophoresis. 2012;33(24):3786–97.
Liu MC, Akle V, Zheng W, Dave JR, Tortella FC, Hayes RL, et al. Comparing calpain- and caspase-3-mediated degradation patterns in traumatic brain injury by differential proteome analysis. Biochem J. 2006;394(3):715–25.
Gyorgy AB, Walker J, Wingo D, Eidelman O, Pollard HB, Molnar A, et al. Reverse phase protein microarray technology in traumatic brain injury. J Neurosci Methods. 2010;192(1):96–101.
Ding Q, Wu Z, Guo Y, Zhao C, Jia Y, Kong F, et al. Proteome analysis of up-regulated proteins in the rat spinal cord induced by transection injury. Proteomics. 2006;6(2):505–18.
Kang S, So H, Moon Y, Kim C. Proteomic analysis of injured spinal cord tissue proteins using 2-DE and MALDI-TOF MS. Proteomics. 2006;6(9):2797–812.
Tsai M, Shen L, Kuo H, Cheng H, Chak K. Involvement of acidic fibroblast growth factor in spinal cord injury repair processes revealed by a proteomics approach. Mol Cell Proteomics. 2008;7(9):1668–87.
Yan X, Liu J, Luo Z, Ding Q, Mao X, Yan M, et al. Proteomic profiling of proteins in rat spinal cord induced by contusion injury. Neurochem Int. 2010;56(8):971–83.
Liu W, Shang F, Xu Y, Belegu V, Xia L, Zhao W, et al. eIF5A1/RhoGDIα pathway: a novel therapeutic target for treatment of spinal cord injury identified by a proteomics approach. Sci Rep. 2015;5:16911.
Chen A, Sun S, Ravikumar R, Visavadiya N, Springer J. Differential proteomic analysis of acute contusive spinal cord injury in rats using iTRAQ reagent labeling and LC–MS/MS. Neurochem Res. 2013;38(11):2247–55.
Moghieb A, Bramlett H, Das J, Yang Z, Selig T, Yost R, et al. Differential neuroproteomic and systems biology analysis of spinal cord injury. Mol Cell Proteomics. 2016;15(7):2379–95.
Devaux S, Cizkova D, Quanico J, Franck J, Nataf S, Pays L et al. Proteomic analysis of the spatio-temporal based molecular kinetics of acute spinal cord injury identifies a time- and segment-specific window for effective tissue repair. Mol Cell Proteomics. 2016;15(8):2641–70.
McDonnell L, Heeren R. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26(4):606–43.
Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60(20):2299–301.
Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, et al. Protein and polymer analyses up tom/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2(8):151–3.
Andersson M, Groseclose M, Deutch A, Caprioli R. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods. 2008;5(1):101–8.
Mainini V, Bovo G, Chinello C, Gianazza E, Grasso M, Cattoretti G, et al. Detection of high molecular weight proteins by MALDI imaging mass spectrometry. Mol Biosyst. 2013;9(6):1101–7.
Clemis E, Smith D, Camenzind A, Danell R, Parker C, Borchers C. Quantitation of spatially-localized proteins in tissue samples using MALDI-MRM imaging. Anal Chem. 2012;84(8):3514–22.
Franck J, Arafah K, Elayed M, Bonnel D, Vergara D, Jacquet A, et al. Maldi imaging mass spectrometry: state of the art technology in clinical proteomics. Mol Cell Proteomics. 2009;8(9):2023–33.
Norris J, Caprioli R. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113(4):2309–42.
Amstalden van Hove E, Smith D, Heeren R. A concise review of mass spectrometry imaging. J Chromatogr A. 2010;1217(25):3946–54.
Sugiura Y, Shimma S, Setou M. Two-step matrix application technique to improve ionization efficiency for matrix-assisted laser desorption/ionization in imaging mass spectrometry. Anal Chem. 2006;78(24):8227–35.
Chaurand P, Caprioli R. Direct profiling and imaging of peptides and proteins from mammalian cells and tissue sections by mass spectrometry. Electrophoresis. 2002;23(18):3125–35.
Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom. 2007;42(2):254–62.
Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proc Natl Acad Sci. 2008;105(47):18126–31.
Taverna D, Norris JL, Caprioli RM. Histology-directed microwave assisted enzymatic protein digestion for MALDI MS analysis of mammalian tissue. Anal Chem. 2015;87(1):670–6.
Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 2010;110(5):3237–77.
Ceuppens R, Dumont D, Brussel LV, Plas BVD, Daniels R, Noben J-P, et al. Direct profiling of myelinated and demyelinated regions in mouse brain by imaging mass spectrometry. Int J Mass Spectrom. 2007;260(2–3):185–94.
Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J Am Soc Mass Spectrom. 2005;16(7):1093–9.
Roux A, Muller L, Jackson SN, Post J, Baldwin K, Hoffer B, et al. Mass spectrometry imaging of rat brain lipid profile changes over time following traumatic brain injury. J Neurosci Methods. 2016;272:19–32.
Woods AS, Colsch B, Jackson SN, Post J, Baldwin K, Roux A, et al. Gangliosides and ceramides change in a mouse model of blast induced traumatic brain injury. ACS Chem Neurosci. 2013;4(4):594–600.
Caughlin S, Hepburn JD, Park DH, Jurcic K, Yeung KK-C, Cechetto DF et al. Increased expression of simple ganglioside species GM2 and GM3 detected by MALDI imaging mass spectrometry in a combined rat model of Aβ toxicity and stroke. PLoS One. 2015;10(6).
Whitehead SN, Chan KHN, Gangaraju S, Slinn J, Li J, Hou ST. Imaging mass spectrometry detection of gangliosides species in the mouse brain following transient focal cerebral ischemia and long-term recovery. PLoS One. 2011;6(6).
Aichler M, Walch A. MALDI imaging mass spectrometry: current frontiers and perspectives in pathology research and practice. Lab Invest. 2015;95(4):422–31.
Grant S. Systems biology in neuroscience: bridging genes to cognition. Curr Opin Neurobiol. 2003;13(5):577–82.
Husi H, Grant SG. Proteomics of the nervous system. Trends Neurosci. 2001;24(5):259–66.
Li C, Chen L, Aihara K. A systems biology perspective on signal processing in genetic network motifs [life sciences]. IEEE Signal Process Mag. 2007;24(2):136–47.
Choudhary J, Grant SGN. Proteomics in postgenomic neuroscience: the end of the beginning. Nat Neurosci. 2004;7(5):440–5.
Beltrao P, Kiel C, Serrano L. Structures in systems biology. Curr Opin Struct Biol. 2007;17(3):378–84.
Feala JD, Abdulhameed MDM, Yu C, Dutta B, Yu X, Schmid K, et al. Systems biology approaches for discovering biomarkers for traumatic brain injury. J Neurotrauma. 2013;30(13):1101–16.
Didangelos A, Puglia M, Iberl M, Sanchez-Bellot C, Roschitzki B, Bradbury E. High-throughput proteomics reveal alarmins as amplifiers of tissue pathology and inflammation after spinal cord injury. Sci Rep. 2016;6:21607.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
George Anis Sarkis, Manasi D. Mangaonkar, Ahmed Moghieb, Brian Lelling, Michael Guertin, Hamad Yadikar, Zhihui Yang, Firas Kobeissy, and Kevin K.W. Wang declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Neurotrauma
George Anis Sarkis, Manasi D. Mangaonkar, Ahmed Moghieb, Firas Kobeissy and Kevin K. W. Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Sarkis, G.A., Mangaonkar, M.D., Moghieb, A. et al. The Application of Proteomics to Traumatic Brain and Spinal Cord Injuries. Curr Neurol Neurosci Rep 17, 23 (2017). https://doi.org/10.1007/s11910-017-0736-z
Published:
DOI: https://doi.org/10.1007/s11910-017-0736-z