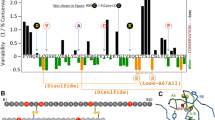
Abstract
Purpose of Review
Diabetes mellitus (DM) due to toxic misfolding of proinsulin variants provides a monogenic model of endoplasmic reticulum (ER) stress. The mutant proinsulin syndrome (also designated MIDY; Mutant INS-gene-induced Diabetes of Youth or Maturity-onset diabetes of the young 10 (MODY10)) ordinarily presents as permanent neonatal-onset DM, but specific amino-acid substitutions may also present later in childhood or adolescence. This review highlights structural mechanisms of proinsulin folding as inferred from phenotype-genotype relationships.
Recent Findings
MIDY mutations most commonly add or remove a cysteine, leading to a variant polypeptide containing an odd number of thiol groups. Such variants are associated with aberrant intermolecular disulfide pairing, ER stress, and neonatal β-cell dysfunction. Non-cysteine-related (NCR) mutations (occurring in both the B and A domains of proinsulin) define distinct determinants of foldability and vary in severity. The range of ages of onset, therefore, reflects a “molecular rheostat” connecting protein biophysics to quality-control ER checkpoints. Because in most mammalian cell lines even wild-type proinsulin exhibits limited folding efficiency, molecular barriers to folding uncovered by NCR MIDY mutations may pertain to β-cell dysfunction in non-syndromic type 2 DM due to INS-gene overexpression in the face of peripheral insulin resistance.
Summary
Recent studies of MIDY mutations and related NCR variants, combining molecular and cell-based approaches, suggest that proinsulin has evolved at the edge of non-foldability. Chemical protein synthesis promises to enable comparative studies of “non-foldable” proinsulin variants to define key steps in wild-type biosynthesis. Such studies may create opportunities for novel therapeutic approaches to non-syndromic type 2 DM.

Similar content being viewed by others
Notes
Abbreviations. DM, diabetes mellitus; ER, endoplasmic reticulum; MIDY, mutant INS-gene-induced Diabetes of Youth; MODY, maturity-onset diabetes of the young; NCR, non-cysteine-related; PND, permanent neonatal-onset DM; and UPR, unfolded-protein response. Residues are designated by standard three-letter code. Residue positions in insulin are shown in superscript (chain and residue number); Leu at position 15 of the B chain, for example, is denoted LeuB15. Cystine pairings are identified by brackets; the disulfide pairing between CysB19 and CysA20, for example, is [B19-A20]. Gene names are italicized.
Although insulin chain combination is in general robust to mutations in the A1-A8 α-helix [91, 92], MODY variant GluA4Lys lies on the surface of this helix. Its effect on the foldability of proinsulin may be due to disruption of a salt bridge with Arg89 (in the dibasic CA junction) in a proinsulin folding intermediate; in the solution structure of a proinsulin monomer this salt bridge appears to provide an N-cap of A1-A8 α-helix [48]. LysA4 could introduce electrostatic repulsion within this element and so attenuate nascent helix formation. A structural puzzle is posed by neonatal-onset MIDY mutation ThrA8Ser [50], also on the surface of insulin.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24(9):329–32.
Iwata K, Fujiwara T, Matsuki Y, Akutsu H, Takahashi S, Naiki H, et al. 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci USA. 2006;103(48):18119–24.
Langkilde AE, Morris KL, Serpell LC, Svergun DI, Vestergaard B. The architecture of amyloid-like peptide fibrils revealed by X-ray scattering, diffraction and electron microscopy. Acta Crystallogr Sect D Biol Crystallogr. 2015;71(4):882–95.
Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat Struct Mol Biol. 2006;13(3):202–8.
Carrell RW, Gooptu B. Conformational changes and disease—serpins, prions and Alzheimer’s. Curr Opin Struct Biol. 1998;8(6):799–809.
Sekijima Y, Wiseman RL, Matteson J, Hammarström P, Miller SR, Sawkar AR, et al. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121(1):73–85.
Canet D, Last AM, Tito P, Sunde M, Spencer A, Archer DB, et al. Local cooperativity in the unfolding of an amyloidogenic variant of human lysozyme. Nat Struct Biol. 2002;9(4):308–15.
Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–90.
Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66.
Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68.
Volchuk A, Ron D. The endoplasmic reticulum stress response in the pancreatic β-cell. Diabetes Obes Metab. 2010;12:48–57.
Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87(4):1377–408.
Roos RA. Huntington’s disease: a clinical review. Orphanet J Rare Dis. 2010;5(1):1–8.
Kumar A, Singh A. A review on Alzheimer’s disease pathophysiology and its management: an update. Pharmacol Rep. 2015;67(2):195–203.
•• Støy J, Edghill EL, Flanagan SE, Ye H, Paz VP, Pluzhnikov A, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci USA. 2007;104(38):15040-4 (This study described the first ten patients with the mutant proinsulin syndrome and provided insights into their diagnosis and treatment.)
•• Colombo C, Porzio O, Liu M, Massa O, Vasta M, Salardi S, et al. Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. J Clin Investig. 2008;118(6):2148-56. (This study (contemporaneous with ref. 15) reported seven additional heterozygous INS mutations causing permanent neonatal DM.)
Steiner D, Clark J, Nolan C, Rubenstein A, Margoliash E, Aten B, et al. Proinsulin and the biosynthesis of insulin. Recent Prog Horm Res. 1969;25:207.
Dodson G, Steiner D. The role of assembly in insulin’s biosynthesis. Curr Opin Struct Biol. 1998;8(2):189–94.
Molven A, Ringdal M, Nordbø AM, Ræder H, Støy J, Lipkind GM, et al. Mutations in the insulin gene can cause MODY and autoantibody-negative type 1 diabetes. Diabetes. 2008;57(4):1131–5.
Edghill EL, Flanagan SE, Patch A-M, Boustred C, Parrish A, Shields B, et al. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes. 2008;57(4):1034–42.
Weiss MA. Diabetes mellitus due to the toxic misfolding of proinsulin variants. FEBS Lett. 2013;587(13):1942–50.
Dhayalan B, Chatterjee D, Chen Y-S, Weiss MA. Structural lessons from the mutant proinsulin syndrome. Front Endocrinol. 2021;12:754693.
Ron D. Proteotoxicity in the endoplasmic reticulum: lessons from the Akita diabetic mouse. J Clin Invest. 2002;109:443–5.
Araki E, Oyadomari S, Mori M. Impact of endoplasmic reticulum stress pathway on pancreatic β-cells and diabetes mellitus. Exp Biol Med. 2003;228(10):1213–7.
Zuber C, Fan J-Y, Guhl B, Roth J. Misfolded proinsulin accumulates in expanded pre-Golgi intermediates and endoplasmic reticulum subdomains in pancreatic beta cells of Akita mice. FASEB J. 2004;18(7):917–9.
Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin maturation, misfolding, and proteotoxicity. Proc Natl Acad Sci USA. 2007;104(40):15841–6.
• Shrestha N, De Franco E, Arvan P, Cnop M. Pathological β-cell endoplasmic reticulum stress in type 2 diabetes: current evidence. Front Endocrinol. 2021;12:650158. (This review highlights the role of the unfolded protein response to the maintainence of ER homeostasis, β-cell function and survival.)
Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529(7586):326–35.
Yang Y, Chan L. Monogenic diabetes: what it teaches us on the common forms of type 1 and type 2 diabetes. Endocr Rev. 2016;37(3):190–222.
Riddle MC, Philipson LH, Rich SS, Carlsson A, Franks PW, Greeley SAW, et al. Monogenic diabetes: from genetic insights to population-based precision in care. Reflections From a Diabetes Care Editors’ Expert Forum. Diabetes Care. 2020;43(12):3117–28.
Nkonge KM, Nkonge DK, Nkonge TN. The epidemiology, molecular pathogenesis, diagnosis, and treatment of maturity-onset diabetes of the young (MODY). Clin Diabetes Endocrinol. 2020;6(1):1–10.
Sousa M, Bruges-Armas J. Monogenic diabetes: genetics and relevance on diabetes mellitus personalized medicine. Curr Diabetes Rev. 2020;16(8):807–19.
Slingerland AS, Hattersley AT. Mutations in the Kir6. 2 subunit of the KATP channel and permanent neonatal diabetes: new insights and new treatment. Ann Med. 2005;37(3):186–95.
Babenko AP, Polak M, Cavé H, Busiah K, Czernichow P, Scharfmann R, et al. Activating mutations in the ABCC8 gene in neonatal diabetes mellitus. N Engl J Med. 2006;355(5):456–66.
Elhaji YA, Hui WuJ, Gottlieb B, Beitel LK, Alvarado C, Batist G, et al. An examination of how different mutations at arginine 855 of the androgen receptor result in different androgen insensitivity phenotypes. Mol Endocrinol. 2004;18(8):1876–86.
Varho TT, Alajoki LE, Posti KM, Korhonen TT, Renlund MG, Nyman SR, et al. Phenotypic spectrum of Salla disease, a free sialic acid storage disorder. Pediatr Neurol. 2002;26(4):267–73.
Nishimura G, Haga N, Kitoh H, Tanaka Y, Sonoda T, Kitamura M, et al. The phenotypic spectrum of COL2A1 mutations. Hum Mutat. 2005;26(1):36–43.
Johannesen K, Marini C, Pfeffer S, Møller RS, Dorn T, Niturad CE, et al. Phenotypic spectrum of GABRA1: from generalized epilepsies to severe epileptic encephalopathies. Neurology. 2016;87(11):1140–51.
Noone PG, Knowles MR. ‘CFTR-opathies’: disease phenotypes associated with cystic fibrosis transmembrane regulator gene mutations. Respir Res. 2001;2(6):1–5.
Remerand G, Boespflug-Tanguy O, Tonduti D, Touraine R, Rodriguez D, Curie A, et al. Expanding the phenotypic spectrum of Allan–Herndon–Dudley syndrome in patients with SLC 16A2 mutations. Dev Med Child Neurol. 2019;61(12):1439–47.
Southard-Smith EM, Angrist M, Ellison JS, Agarwala R, Baxevanis AD, Chakravarti A, et al. The Sox10Dom mouse: modeling the genetic variation of Waardenburg-Shah (WS4) syndrome. Genome Res. 1999;9(3):215–25.
Åkerblom HK, Vaarala O, Hyöty H, Ilonen J, Knip M. Environmental factors in the etiology of type 1 diabetes. Am J Med Genet. 2002;115(1):18–29.
Baker EN, Blundell TL, Cutfield JF, Dodson EJ, Dodson GG, Hodgkin DMC, et al. The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos Trans R Soc Lond B Biol Sci. 1988;319(1195):369–456.
Hua QX, Mayer J, Jia W, Zhang J, Weiss MA. The folding nucleus of the insulin superfamily: a flexible peptide model foreshadows the native state. J Biol Chem. 2006;281:28131–42.
Narhi LO, Hua QX, Arakawa T, Fox GM, Tsai L, Rosenfeld R, et al. Role of native disulfide bonds in the structure and activity of insulin-like growth factor 1: genetic models of protein-folding intermediates. Biochemistry. 1993;32:5214–21.
Sieber P, Eisler K, Kamber B, Riniker B, Rittel W, Märki F, et al. Synthesis and biological activity of two disulphide bond isomers of human insulin:[A7-A11, A6-B7-cystine]-and [A6-A7, A11-B7-cystine] insulin (human). Biol Chem. 1978;359(1):113–24.
Hua QX, Jia W, Frank BH, Phillips NB, Weiss MA. A protein caught in a kinetic trap: structures and stabilities of insulin disulfide isomers. Biochemistry. 2002;41:14700–15.
Yang Y, Hua QX, Liu J, Shimizu EH, Choquette MH, Mackin RB, et al. Solution structure of proinsulin: connecting domain flexibility and prohormone processing. J Biol Chem. 2010;285:7847–51.
Steiner DF, Tager HS, Chan SJ, Nanjo K, Sanke T, Rubenstein AH. Lessons learned from molecular biology of insulin-gene mutations. Diabetes Care. 1990;13:600–9.
• Støy J, De Franco E, Ye H, Park S-Y, Bell GI, Hattersley AT. In celebration of a century with insulin–update of insulin gene mutations in diabetes. Mol Metab. 2021:101280. (This recent review provides an update to catalogue of INS mutations and discusses genetic causes and clinical opportunities for diagnosis and treatment.)
Miller JA, Narhi LO, Hua QX, Rosenfeld R, Arakawa T, Rohde M, et al. Oxidative refolding of insulin-like growth factor 1 yields two products of similar thermodynamic stability: a bifurcating protein-folding pathway. Biochemistry. 1993;32:5203–13.
Guo ZY, Qiao ZS, Feng YM. The in vitro oxidative folding of the insulin superfamily. Antioxid Redox Signal. 2008;10:127–40.
Finkelstein AV, Ivankov DN, Garbuzynskiy SO, Galzitskaya OV. Understanding the folding rates and folding nuclei of globular proteins. Curr Protein Peptide Sci. 2007;8(6):521–36.
Haataja L, Manickam N, Soliman A, Tsai B, Liu M, Arvan P. Disulfide mispairing during proinsulin folding in the endoplasmic reticulum. Diabetes. 2016;65(4):1050–60.
Liu M, Weiss MA, Arunagiri A, Yong J, Rege N, Sun J, et al. Biosynthesis, structure, and folding of the insulin precursor protein. Diabetes Obes Metab. 2018;20:28–50.
Herbach N, Rathkolb B, Kemter E, Pichl L, Klaften M, de Angelis MH, et al. Dominant-negative effects of a novel mutated Ins2 allele causes early-onset diabetes and severe β-cell loss in Munich Ins2C95S mutant mice. Diabetes. 2007;56(5):1268–76.
Matthews BW. Structural and genetic analysis of protein stability. Annu Rev Biochem. 1993;62(1):139–60.
Creighton TE. Protein folding: an unfolding story. Curr Biol. 1995;5(4):353–6.
Eriksson AE, Baase WA, Zhang X-J, Heinz DW, Blaber M, Baldwin EP, et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255(5041):178–83.
Mezei M. On predicting foldability of a protein from its sequence. Proteins Struct Funct Bioinform. 2020;88(2):355–65.
Xu J, Baase WA, Baldwin E, Matthews BW. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7:158–77.
Eriksson AE, Baase WA, Wozniak JA, Matthews BW. A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature. 1992;355(6358):371–3.
O’Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250:646–51.
Padmanabhan S, Marqusee S, Ridgeway T, Laue TM, Baldwin RL. Relative helix-forming tendencies of nonpolar amino acids. Nature. 1990;344(6263):268–70.
Ortolani F, Piccinno E, Grasso V, Papadia F, Panzeca R, Cortese C, et al. Diabetes associated with dominant insulin gene mutations: outcome of 24-month, sensor-augmented insulin pump treatment. Acta Diabetol. 2016;53(3):499–501.
• Haataja L, Arunagiri A, Hassan A, Regan K, Tsai B, Dhayalan B, et al. Distinct states of proinsulin misfolding in MIDY. Cell Mol Life Sci. 2021;78:6017–31. (This study highlights the role of unpaired cysteines and proinsulin-mutant proinsulin self-association in the misfolding and ER stress using seven MIDY mutations.)
• Weiss MA, Nakagawa SH, Jia W, Xu B, Hua QX, Chu YC, et al. Protein structure and the spandrels of San Marco: insulin’s receptor-binding surface is buttressed by an invariant leucine essential for protein stability. Biochemistry. 2002;41:809-19. (This paper highlights the essential contribution of LeuA16 to chain-combination efficiency as a molecular “spandrels of San Marco. ”)
•• Liu M, Wan Z-l, Chu Y-C, Aladdin H, Klaproth B, Choquette M, et al. Crystal structure of a “nonfoldable” insulin impaired folding efficiency despite native activity. J Biol Chem. 2009;284(50):35259-72. (This paper demonstrates that substitution LeuA16→Val preserves structure and function but markedly impairs nascent disulfide pairing in proinsulin and blocks insulin chain combination in anticipation of its observation as a recessive mutation in neonatal diabetes.)
Blundell T, Humbel R. Hormone families: pancreatic hormones and homologous growth factors. Nature. 1980;287(5785):781–7.
Menting JG, Yang Y, Chan SJ, Phillips NB, Smith BJ, Whittaker J, et al. Protective hinge in insulin opens to enable its receptor engagement. Proc Natl Acad Sci U S A. 2014;111(33):E3395–404.
Scapin G, Dandey VP, Zhang Z, Prosise W, Hruza A, Kelly T, et al. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature. 2018;556(7699):122–5.
Weis F, Menting JG, Margetts MB, Chan SJ, Xu Y, Tennagels N, et al. The signalling conformation of the insulin receptor ectodomain. Nat Commun. 2018;9(1):1–10.
Gould SJ. The exaptive excellence of spandrels as a term and prototype. Proc Natl Acad Sci USA. 1997;94(20):10750–5.
Li M, Rivière J-B, Polychronakos C. Why all MODY variants are dominantly inherited: a hypothesis. Trends Genet. 2021.
Al-Muhaizea MA, Aldeeb H, Almass R, Jaber H, Binhumaid F, Alquait L, et al. Genetics of ataxia telangiectasia in a highly consanguineous population. Ann Hum Genet. 2021;86(1):34–44.
Doyle AC, Paget S. The adventure of silver blaze: Mary McLaughlin and M. Einisman for the Scotland Yard Bookstore; 1979.
Chan SJ, Cao Q-P, Steiner DF. Evolution of the insulin superfamily: cloning of a hybrid insulin/insulin-like growth factor cDNA from amphioxus. Proc Natl Acad Sci USA. 1990;87(23):9319–23.
Bliss M. The discovery of insulin: twenty-fifth. anniversary. Chicago: University of Chicago Press; 2007. p. 310.
Flier JS, Kahn CR. Insulin: a pacesetter for the shape of modern biomedical science and the Nobel Prize. Mol Metab. 2021:101194.
Luisier S, Avital-Shmilovici M, Weiss MA, Kent SB. Total chemical synthesis of human proinsulin. Chem Commun. 2010;46(43):8177–9.
Kent SB. Novel protein science enabled by total chemical synthesis. Protein Sci. 2019;28(2):313–28.
Avital-Shmilovici M, Whittaker J, Weiss MA, Kent SB. Deciphering a molecular mechanism of neonatal diabetes mellitus by the chemical synthesis of a protein diastereomer,[D-AlaB8] human proinsulin. J Biol Chem. 2014;289(34):23683–92.
Rege NK, Liu M, Yang Y, Dhayalan B, Wickramasinghe NP, Chen Y-S, et al. Evolution of insulin at the edge of foldability and its medical implications. Proc Natl Acad Sci USA. 2020;117(47):29618–28.
Hodish I, Liu M, Rajpal G, Larkin D, Holz RW, Adams A, et al. Misfolded proinsulin affects bystander proinsulin in neonatal diabetes. J Biol Chem. 2010;285(1):685–94.
Sun J, Xiong Y, Li X, Haataja L, Chen W, Mir SA, et al. Role of proinsulin self-association in mutant INS gene–induced diabetes of youth. Diabetes. 2020;69(5):954–64.
Sun J, Cui J, He Q, Chen Z, Arvan P, Liu M. Proinsulin misfolding and endoplasmic reticulum stress during the development and progression of diabetes. Mol Aspects Med. 2015;42:105–18.
Liu M, Hodish I, Haataja L, Lara-Lemus R, Rajpal G, Wright J, et al. Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends Endrocrinol Metab. 2010;21(11):652–9.
• Yong J, Johnson JD, Arvan P, Han J, Kaufman RJ. Therapeutic opportunities for pancreatic β-cell ER stress in diabetes mellitus. Nat Rev Endocrinol. 2021:1–13. (This review highlights β-cell ER stress and evaluates related therapeutic opportunities to maintain functional β-cell mass in non-syndromic type 2 DM.)
Fishman MC. Power of rare diseases: found in translation. Sci Transl Med. 2013;5(201):201ps11-ps11.
Jarosinski MA, Dhayalan B, Chen Y-S, Chatterjee D, Varas N, Weiss MA. Structural principles of insulin formulation and analog design: a century of innovation. Mol Metab. 2021:101325.
Hua QX, Chu YC, Jia W, Phillips NB, Wang RY, Katsoyannis PG, et al. Mechanism of insulin chain combination. Asymmetric roles of A-chain α-helices in disulfide pairing. J Biol Chem. 2002;277:43443–53.
Weiss MA, Wan Z, Zhao M, Chu Y-C, Nakagawa SH, Burke GT, et al. Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor. J Mol Biol. 2002;315(2):103–11.
Granel B, Valleix S, Serratrice J, Cherin P, Texeira A, Disdier P, et al. Lysozyme amyloidosis: report of 4 cases and a review of the literature. Medicine (Baltimore). 2006;85(1):66–73.
Benson MD, Liepnieks J, Uemichi T, Wheeler G, Correa R. Hereditary renal amyloidosis associated with a mutant fibrinogen α–chain. Nat Genet. 1993;3(3):252–5.
Ohashi K. Pathogenesis of β2-microglobulin amyloidosis. Pathol Int. 2001;51(1):1–10.
Benson MD, Uemichi T. Transthyretin amyloidosis. Amyloid. 1996;3(1):44–56.
Merlini G, Comenzo RL, Seldin DC, Wechalekar A, Gertz MA. Immunoglobulin light chain amyloidosis. Expert Rev Hematol. 2014;7(1):143–56.
Prusiner SB. Molecular biology of prion diseases. Science. 1991;252(5012):1515–22.
Revesz T, Holton JL, Lashley T, Plant G, Frangione B, Rostagno A, et al. Genetics and molecular pathogenesis of sporadic and hereditary cerebral amyloid angiopathies. Acta Neuropathol. 2009;118(1):115–30.
Kiuru S. Gelsolin-related familial amyloidosis, Finnish type (FAF), and its variants found worldwide. Amyloid. 1998;5(1):55–66.
Leboulleux S, Baudin E, Travagli JP, Schlumberger M. Medullary thyroid carcinoma. Clin Endocrinol (Oxf). 2004;61(3):299–310.
Röcken C, Peters B, Juenemann G, Saeger W, Klein HU, Huth C, et al. Atrial amyloidosis: an arrhythmogenic substrate for persistent atrial fibrillation. Circulation. 2002;106(16):2091–7.
Sotiropoulos I, Galas M-C, Silva JM, Skoulakis E, Wegmann S, Maina MB, et al. Atypical, non-standard functions of the microtubule associated Tau protein. Acta Neuropathol Commun. 2017;5(1):1–11.
Du K, Sharma M, Lukacs GL. The ΔF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol. 2005;12(1):17–25.
Kereszturi É, Szmola R, Kukor Z, Simon P, Ulrich Weiss F, Lerch MM, et al. Hereditary pancreatitis caused by mutation-induced misfolding of human cationic trypsinogen: a novel disease mechanism. Hum Mutat. 2009;30(4):575–82.
Tagliavacca L, Wang Q, Kaufman RJ. ATP-dependent dissociation of non-disulfide-linked aggregates of coagulation factor VIII is a rate-limiting step for secretion. Biochemistry. 2000;39(8):1973–81.
Lang AE, Lozano AM. Parkinson’s disease. N Engl J Med. 1998;339(16):1130–43.
Dhayalan B, Glidden MD, Zaykov A, Chen Y-S, Yang Y, Phillips NB, et al. Peptide Model of the Mutant Proinsulin Syndrome. I. Design and Clinical Correlation. Front Endocrinol. 2022;13:821069
Yang Y, Glidden MD, Dhayalan B, Zaykov A, Chen Y-S, Wickramasinghe NP, et al. Peptide Model of the Mutant Proinsulin Syndrome. II. Nascent NMR Structure and Biophysical Correlation. Front Endocrinol. 2022;13:821091
Acknowledgements
The authors thank P. Arvan, T.L. Blundell, D. Chatterjee, Y.S. Chan, M. Jarosinski, S.B. Kent, F. Ismail-Beigi, M. Liu, N.B. Phillips, J. Racca, and Y. Yang for helpful discussion. We thank L. Liu, P. Arvan, G.I. Bell, L. Philipson, and E. De Franco for communication of results prior to publication. M.A.W. is grateful to the late G.G. Dodson, P.G. Katsoyannis, and D.F. Steiner for their encouragement in the early years of this research program.
Funding
This work was supported in part by a grant from the National Institutes of Health (R01 DK040949). BD is supported in part by a grant from the Diabetes Research Connection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Pediatric Type 2 and Monogenic Diabetes
Rights and permissions
About this article
Cite this article
Dhayalan, B., Weiss, M.A. Diabetes-Associated Mutations in Proinsulin Provide a “Molecular Rheostat” of Nascent Foldability. Curr Diab Rep 22, 85–94 (2022). https://doi.org/10.1007/s11892-022-01447-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11892-022-01447-2