Abstract
Detection of molecular markers for colorectal neoplasia in feces has the potential to improve performance of simple noninvasive screening tests for colorectal cancer. Most research has explored the value of DNA-based, RNA-based, and protein-based markers. In all cases there has been a trend to move from a single marker to a panel of markers to improve sensitivity. Unfortunately, no type of molecular marker has proved specific for neoplasia. DNA tests have been improved by combining mutation detection with assessment of DNA integrity plus epigenetic markers of neoplasia. RNA-based approaches are just beginning to explore the full power of transcriptomics. So far, no protein-based fecal test has proved better than fecal immunochemical tests for hemoglobin. Finally, no marker or panel of markers has yet been developed to the point where it has been evaluated in large unbiased population studies to assess performance across all stages of neoplasia and in all practical environments.
Similar content being viewed by others
Introduction
Progressive genomic instability in the colorectum gives rise to colorectal cancer (CRC) due to the accumulation of genetic alterations (gene mutations, gene amplification, etc.) and epigenetic alterations (aberrant DNA methylation, chromatin modifications, etc.) that transform epithelial cells into carcinoma cells. Altered function of proto-oncogenes and tumor suppressor genes results in disturbances of biological processes such as DNA repair, proliferation, and apoptosis. Several different molecular pathways of oncogenesis have been defined [1•, 2]. Our understanding of the underlying and associated molecular changes is increasing; specific changes at the DNA, RNA, and protein levels are being discovered as well as metabolic and other changes. These “molecular” changes have the potential to serve as biomarkers for neoplasia and may be useful for screening, diagnosis, prognostication, predicting responses to therapy, and detecting recurrence provided that they can be measured in biological materials such as blood or feces.
This review aims to provide an update on the concepts underlying the use of molecular biomarkers for screening, how new biomarkers can be evaluated, and which show promise for use in screening. We will not consider tests for fecal blood in this review, even though some specifically target the hemoglobin molecule (ie, fecal immunochemical tests, FIT [3•]) nor will we consider blood-based molecular tests which are fast becoming an increasingly active area of research.
Proven Simple Screening Tests
Multiple population-based randomized trials have proved that guaiac-based fecal occult blood tests (GFOBT) detect early-stage disease and result in a reduced mortality from and incidence of CRC [4, 5]. In other words, early detection of lesions with a “bleeding phenotype” is beneficial in that one can be confident that treatment (surgery, polypectomy, etc.) will result in worthwhile gains.
A single targeted population-based randomized trial has proved that flexible sigmoidoscopy detects early-stage disease and results in a reduced mortality from and incidence of CRC [6••]. Detection by endoscopic methods requires only that a lesion be visualizable and since such is independent of the bleeding phenotype, this trial indicates that treatment benefit for early neoplastic lesions is probably independent of tumor phenotype or genotype. This is a crucial point because expression of a molecular biomarker by a tumor might reflect a certain biological behavior that reflects natural history or is related to or determines response to therapy. For example, a lesion might grow so slowly as to not even need treatment within a lifetime or, conversely, it might be so aggressive as to render curative treatment pointless.
Evaluating New Screening Biomarkers
There are relatively few practical guidelines on how best to compare “new” with proven screening tests and it is not the purpose of this review to critique these strategies but it is clear that new tests cannot be initially subject to randomized controlled trials (RCTs) with mortality or incidence as the end point. Strategically, it is expedient to evaluate a biomarker’s diagnostic performance relative to a proven comparator screening test in highly selected cases with cancer [7]. But evaluation of test accuracy must be backed up by programmatic evaluation in the screening context where prevalence is low and non-neoplastic lesions are present [8] such that we know how the test performs across the spectrum of prevalent neoplastic lesions. Such is essential for a new test to be adopted for widespread use.
A phased evaluation of a new biomarker is recommended [8]. Initial evaluation of biomarkers will usually be undertaken in small numbers of highly selected cases, often retrospective (such as those with symptomatic cancer versus normal controls) and using colonoscopy as the reference standard [9]. If promising, evaluation should proceed to comparison with a proven screening test such as an FOBT (ideally a FIT [3•]) but such need not include cancer-specific mortality as an end point provided that the comparator test’s effect on population mortality has been proven by a population randomized controlled trial [7]. Guaiac-FOBT effectiveness represents the minimum standard to be achieved by a new test since GFOBT are effective (albeit limited) and inexpensive. Finally, a prospective evaluation of performance across the continuum of neoplastic lesions from adenoma to cancer should be undertaken in large unselected typical screening populations.
Such comparative studies focus initially on relative detection, but screening is about more than detection; it is about success when applied to many healthy people where disease prevalence is low and acceptability of the test is crucial. Demonstration of adequate accuracy in prescreening phases justifies progression to mass-population studies that address programmatic outcomes on an intention-to-screen basis followed by evaluation of cost-effectiveness in ongoing screening.
It will be apparent from the following discussions that fecal molecular tests have not yet progressed to evaluation by randomized fashion in large unselected populations.
How Might Molecular Markers Improve Screening Outcomes?
Screening by existing proven methods has disadvantages one way or another. Endoscopic methods are invasive and inconvenient and when used as the primary screening tool are used on many people who will prove to have no neoplastic lesions in the colon. FOBT methods, while simple and increasing the likelihood of neoplasia being present when positive, are not specific for neoplasia, and have somewhat limited sensitivity for cancer and especially adenomas [9, 10], although FIT are a major improvement over GFOBT [3•].
It would be useful to have simple and acceptable noninvasive screening tests that are more specific for neoplasia, ie, have a lower false-positive rate. If so, this will reduce costs of diagnostic follow-up, workloads, and overall program cost.
It would also be valuable if simple and acceptable noninvasive screening tests were more sensitive for curable cancer and more advanced adenomas, ie, return a higher true-positive rate. This would improve impact on mortality and incidence.
While it is known that limitations of existing simple tests, namely FOBT, include lack of specificity for neoplasia and variation between tumors as to whether they bleed or not [11], it needs to be shown, if simple molecular tests are to replace FOBT, that they do indeed point to molecular phenomena that are either more predictable than bleeding and/or more characteristic of neoplasia.
Finally, sensitivity and specificity are both important and improving one at the expense of the other does not necessarily provide an advantage.
Detection of Neoplasia Using Biomarkers
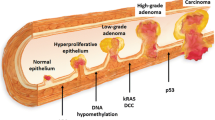
Colorectal oncogenesis follows several genetic pathways [1•, 2]. The process is protracted and takes years, perhaps decades, from the initial event that initiates oncogenesis until sufficient molecular events accumulate to change cell behavior and result in the invasive phenotype (ie, cancer) [1•, 12]. This sequence underlies the phenotypic progression known as the adenoma-carcinoma sequence. The stage of neoplasia at which a biomarker becomes expressed and detectable (ie, “positive”) is crucial to what we hope molecular biomarkers will detect in the screening context. A positive biomarker initiates diagnostic confirmation by colonoscopy.
Figure 1 provides a conceptual presentation of the most advanced stage of colorectal neoplasia reached by the time of death (“life-time state”) for a typical Western population [11••]. Curable neoplastic lesions are the obvious target. Curable cancer (point C in the process in Fig. 1) is a key target, as the GFOBT RCTs showed that detection of curable cancer (ie, downstaging of cancer) leads to reduction in mortality [4]. Detection and removal of adenomas (particularly advanced adenomas, point B in Fig. 1) is also a worthwhile target as the flexible sigmoidoscopy RCT showed a reduction in incidence as did one GFOBT RCT [5, 6••]. These studies support the recent US multi-society guidelines that recommended that screening target not just early-stage cancer but also “advanced” adenomas [13••]. Advanced adenomas are defined as those of >9 mm, those with villous change, or those showing high-grade dysplasia [14].
It can be seen that different biomarkers might characterize different stages of oncogenesis and so vary in their usefulness.
Small, tubular, low-grade adenomas (point A in Fig. 1) are more common than large, high-grade, or villous lesions. Detection of early, small adenomas might not be useful in that they pose little risk for CRC [15]—detection of inconsequential neoplastic lesions is referred to as over-diagnosis and is a major issue for prostate cancer screening [16]. Detection of incurable neoplasia (biomarker point D in Fig. 1), likewise, would not lead to benefit.
In other words, there is a gradation of probability of progression from adenoma to cancer, with the more advanced lesions more likely to progress to cancer; biomarkers for adenomas will vary in their capacity to predict likelihood of progression to cancer according to the stage detected, as not all small adenomas progress to advanced adenomas [14, 17, 18], and test sensitivity.
It also needs to be noted that with the different molecular pathways of oncogenesis, some molecular markers might be pathway specific. So, to reliably detect neoplasms, a panel of molecular markers might be needed.
Biomarkers in Feces
Apart from these biological considerations with regard to different stages of and pathways to cancer, we also need to consider the way in which a biomarker gets into the biological sample in which we want to measure it such as feces, and whether it remains detectable. This is important because most biomarkers are first “discovered” in neoplastic tissues but will be sought in biological samples where their chemical nature may be different.
Feces is comprised of water, undigested food, bacterial biomass, shed cells, fragments of cells, and other products from cells and vessels (blood and lymphatics). A clinically useful test for a biomarker must be able to detect a molecule in this complex and highly destructive mix that contains many enzymes and other substances that will change the characteristics of a marker, often by degradation.
The evidence indicates that neoplasms are associated with “leakage” of cellular and vascular materials as well as shedding of cells into feces. Colonocytes are continuously shed into the fecal stream [19••]. Markers can be classified as those that leak through neoplasms, are secreted by neoplasms, or arise by exfoliation [20]. To these we should add shedding of cell fragments by events such as apoptosis and release of tiny membranous vesicles (termed exosomes) [21]. Biomarkers might arise from the neoplasm itself, reflect a cellular response to the neoplasm, or simply result from its physical presence (eg, leakage of plasma proteins). Whatever the case, the biomarker needs to be stable to ensure reliable detection. This might require a sample such as feces to be collected under stringent conditions so as to ensure its usefulness. This has proved to be crucial even for DNA, which is known to be more stable than protein or RNA in biological samples. A device appropriate for ensuring that fecal DNA is of adequate quality has been developed [22], but this is costly and logistically involved.
Ultimately, stability of a biomarker in feces and simplicity of collection of a stable sample will be crucial determinants of the practical usefulness of a biomarker.
Molecular Markers in Feces
As indicated above, it is known that biomarkers may gain access to feces by many routes. It is also known that neoplastic cells are less subject to apoptotic breakdown and represent a proportionally higher fraction of fecal epithelial cells when neoplasia is present [23]. Potential markers could fall anywhere along the “omics” flow of information processing relating to cell phenotype and behavior:
-
Genomics: DNA, reflecting genetic (eg, mutations) and epigenetic changes (eg, aberrant methylation) that are characteristic of colorectal oncogenesis
-
Transcriptomics: mRNA and microRNA, reflecting expression patterns characteristic of neoplasia
-
Proteomics: Proteins, reflecting abnormalities dependent on translational or post-translational processing
-
Metabolomics: Other biochemical events characteristic of neoplasia.
Biochemical methods such as mutation analysis, next-generation sequencing, microarrays, and proteomics may all provide an option for detecting markers.
It is important to note that molecular markers in feces potentially arise from any region of the aerodigestive system. Ideally, one would use a marker that has selectivity for colorectal neoplasia. A marker not specific for a given organ-cancer raises the possibility of needing to pursue the possibility of cancer anywhere in the aerodigestive tract. This would be costly and of questionable benefit.
Fecal DNA Markers
DNA Markers in Feces: Pros and Cons
In a recent review, it has been pointed out that most studies of stool-based DNA biomarkers have focused on the detection of aberrant DNA originating from cancers [19••]. Several biological mechanisms help to increase the potential to specifically detect tumor DNA in stool. Whereas DNA in normal cells is degraded upon shedding by rapid induction of apoptosis, shed tumor cells may escape from apoptosis with reduced loss of DNA integrity because apoptosis is reduced in neoplasms [23]. Thus, a simple general measure of DNA integrity may be useful.
Genetic markers may, alternatively, be very specific. But, many different point mutations characterize colorectal oncogenesis and while a given gene might be a common target for mutations, many different gene probes will be needed to detect all the relevant mutations. Furthermore, the mere presence of an oncogenic mutation in a cell does not guarantee progression to cancer [1•]. Consequently, mutation detection is biologically limited, perhaps more so than is detection of a bleeding phenotype using an FOBT.
Next to genetic alterations such as mutations, epigenetic changes play an important role in deregulating gene expression in CRC [2]. Hypermethylation of the promoter region of a gene, which can cause silencing of tumor suppressor genes in many cancers, is a well studied example. Many genes have been reported to be hypermethylated in colorectal cancer [24], and these alterations appear to be early events in carcinogenesis [19••]. The latest developments in stool-based DNA tests take advantage of both genetic (precise and global) and epigenetic changes.
DNA Biomarker Evaluation
Bosch et al. [19••] provide a comprehensive and thorough review of stool-based DNA markers and the present review will focus on the key issues arising from the many studies undertaken. Bosch et al. found 19 papers looking at multiple biomarkers in stool and 18 evaluating “single” (or single class) gene markers [19••]. For single markers, mutations in K-ras and APC were initially studied and eventually discarded as being useful in themselves. Most studies were small, involved highly selected clinical (rather than screen-detected) cohorts, and cases with adenomas were generally not included; as a generalization, sensitivities for cancer ranged from around 40% to almost 90%.
Investigators progressed to include methylation and other markers. A range of genes has been tested for methylation—SFRP2, TFPI2, GATA4, NDRG4, OSMR, and vimentin—with no one marker emerging as obviously the best [19••]. The DNA integrity assay for long-chain DNA (the DIA assay) [23] has also been included [25•]. As a generalization, specificities ranged from 80% upwards for these markers, making it clear that such molecular tests did not guarantee specificity for neoplasia.
In parallel, investigators have pursued the value of multiple markers, evaluated as a panel. Combinations of mutation analysis of several genes with and without markers of MSI (microsatellite instability), methylation, and or DIA have been evaluated, again mainly in highly selected clinical cohorts of patients with cancer. As a generalization, detection was somewhat improved and specificity tended to be in the range of 90% to 95% [19••].
A large study promised much for DNA panels [26]. The detection rate of cancer using a panel of markers based on key tumor suppressor and oncogenes plus the DIA assay and MSI, was 52% sensitive for cancer compared to 13% using Hemoccult II GFOBT (P = 0.003). The sensitivity of both the panel and the GFOBT for detecting cancer or advanced adenoma was poor, however: 18.2% for the DNA panel compared to 10.8% for Hemoccult II (P = .001). The performance of both tests in detecting advanced adenomas was similarly disappointing (15.1% vs 10.7%, respectively). In patients with negative colonoscopy, 5.6% tested positive on fecal DNA (94.4% specific), compared to 4.8% with Hemoccult II (95.2% specific). The study has been criticized because of the uncharacteristically low sensitivity for cancer of Hemoccult II.
Clearly, even a comprehensive DNA panel is biologically limited. Furthermore, the specificity of fecal DNA was no better than that of Hemoccult II. This might seem surprising, but the adenoma-carcinoma sequence is propagated through an accumulation of genomic events. Each event of itself is not capable of driving the oncogenic pathway to the point of colonoscopically detectable neoplasia.
A similarly large study was subsequently reported using some technological advances of the same panel [27••]. This time, the panel detected only 25% of cancers (less than did the GFOBT Hemoccult) and 17% of adenomas greater than 1 cm, with a specificity of 96%.
Inclusion of vimentin methylation improved detection. Ahlquist et al. [27••] adapted the panel to comprise vimentin together with mutation analysis of K-ras and APC genes and detected 58% of cancers. Itzkowitz et al. [28•] combined vimentin with DIA to achieve 86% sensitivity but specificity was inadequate: the two studies returned specificities of 84% and 73%, respectively.
Table 1 summaries the larger DNA studies (more than 200 cases/controls) published since 2003.
Conclusions
DNA tests have great potential but as yet the ideal panel is far from clear. The technology continues to evolve. Thorough evaluation of such markers relative to different molecular pathways of oncogenesis remains to be undertaken. Costs of molecular tests remain high relative to FOBT although with advances in technology this seems likely to change. Unbiased mass population-based studies have not yet been undertaken.
Fecal RNA Markers
RNA Markers in Feces: Pros and Cons
Tumor-derived RNA seems likely to get into stool by the same processes as DNA. In addition, RNA might gain access if cells secrete tiny membranous vesicles (termed exosomes) that carry RNA species from the cell of origin [21]. Various types of RNA, not just mRNA (which code for proteins) but also microRNA (which are noncoding but greatly influence gene expression), will gain access to feces by these mechanisms. MicroRNAs have potential to be biomarkers for cancers [31].
We know less about the value of RNA markers. A common approach has been to “discover” potential RNA-based markers by examining gene expression profiles, called transcriptomics, in neoplastic compared to normal tissue and then pursuing those markers that appear to differentiate neoplasia from non-neoplasia [8]. Underlying this is the concept that gene expression patterns are unique for neoplasia (and ideally for neoplasia within a given organ) and that the differences between neoplasia and non-neoplasia are greater than variations within non-neoplasia (eg, along the length of the colon [32•] or variations related to diet). Measuring patterns of expression of large numbers of mRNA markers is becoming commonplace in biological research, although initial studies have focused on a single marker or small panel when searching for fecal markers [19••].
Unfortunately, RNA is less stable than DNA and assay in feces creates major methodological challenges for preservation of samples and RNA stability.
RNA Biomarker Evaluation
The potential for stool RNA markers to be informative has been reviewed by Bosch et al. [19••]. Studies have focused principally on a single or small panel of markers.
To date the most studied mRNA marker in stool has been PTGS2 (Cox-2, prostaglandin-synthase 2), but all studies have been restricted to highly selected cases with cancer. Table 2 shows that sensitivities have ranged from 50% to 90%, with specificities of 93% or higher [33–36]. Much larger cohorts that include disease controls are required if we are to assess whether this biomarker realizes its potential for use in a typical screening population.
Using larger panels of markers, for example, oligonucleotide microarrays, cancer-specific gene expression profiles can be built and used to discriminate between cancer and non-cancer. Using this approach, one study revealed that a panel of nine oligonucleotide probes (PAP, REG1A, DPEP1, SEPP1, RPL27A, ATP1B1, EEF1A1, SFN, and RPS11) gave a sensitivity of 78% for CRC, with a specificity of 100% [37]. However, they used fecal colonocytes as the specimen for assay and applicability to stools is unclear.
Conclusions
Technological developments will have a big influence on whether this approach based on RNA, whether mRNA or microRNA, bears fruit. A panel of biomarkers seems likely to be needed. For now, we are far from a feasible test for clinical practice.
Fecal Protein Markers
Protein Markers in Feces: Pros and Cons
The detection of proteins in feces is relatively easy since the protein of interest is often readily detected in small sample volumes using inexpensive technologies. FIT are just one example where simple qualitative or quantitative methods for detection can be developed and provided at a low cost. If new markers are shown to be stable in stool, then simple test platforms such as membrane lateral flow immunochemical devices or quantitative latex-agglutination can be developed to provide simple tests for mass screening.
While some proteins are quite stable in feces, many undergo degradation by proteolytic enzymes. Glycoproteins are particularly subject to attack by glycosidases. Mass spectrometry may well provide means for detecting degraded markers and so “proteomics” might well deliver useful biomarkers.
Protein Biomarker Evaluation
To date a moderately large number of markers have been studied. Fifteen studies have been reviewed in depth [19••]. Apart from immunochemical assays for human hemoglobin itself, none has established a novel marker yet, although most studies have been simple cohort studies with few disease controls.
Markers range from proteins derived from blood or plasma (eg, hemoglobin, calprotectin, haptoglobin) to possibly tumor-derived markers such as M2-PK–pyruvate kinase type M2, S100A12–S100 calcium-binding protein A12, and TIMP-1–TIMP metallopeptidase inhibitor 1 [19••].
Table 3 summarizes those papers that have included two markers of interest, M2-PK and S100A12, that seem derived from tumors. Tumor M2-PK appears to have considerable potential [39, 40], with sensitivity for cancer in a selected cohort of 78% to 85% and for adenomas in general of 23% to 38%. Combining S100A12 with hemoglobin, haptoglobin, and TIMP-1 in a study of over 500 patients shows sensitivity for cancer around 85% with better than 95% specificity [41].
Markers arising from leakage of blood have shown some ability to discriminate neoplasia from non-neoplasia but these have not replaced FIT in practice. These include the proteins haptoglobin [42, 43] and calprotectin, although the latter is highly sensitive for inflammatory bowel disease [44].
Conclusions
Some of these show promise, but as with other biomarkers, until they are compared to a proven screening test such as GFOBT and evaluated in large typical screening populations, the potential usefulness remains uncertain. Extensive evaluation in disease controls is also needed because markers evaluated so far are not necessarily tumor-specific markers.
General Conclusions
The search for molecular biomarkers useful for screening for colorectal cancer has not yet led to a simple test that can replace FOBT. While good sensitivity has been achieved for cancer, sensitivity for adenomas has not been adequately explored. Furthermore, molecular tests are not proving to be any more specific for neoplasia than are FOBT, especially as tests are configured to optimize sensitivity. DNA tests have been improved by combining mutation detection with assessment of DNA integrity plus epigenetic markers of neoplasia. RNA-based approaches are just beginning to explore the full power of transcriptomics. So far, no protein-based fecal test has proved better than fecal immunochemical tests for hemoglobin. It seems likely that a panel of markers will be required to ensure that the various molecular pathways of oncogenesis and the different patterns of gene expression are adequately covered.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
•Grady WM, Carethers JM: Gastroenterology 2008;135:1079–99. Genomic and epigenetic instability in colorectal cancer pathogenesis. This is a useful summary of molecular pathways of oncogenesis.
Weisenberger DJ, Siegmund KD, Campan M, et al.: CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787–93.
•Young GP, Cole S: Which fecal occult blood test is best to screen for colorectal cancer? Nature Clinical Practice Gastroenterology and Hepatology, 2009;6:140–141. This is a succinct review of why fecal immunochemical tests should replace guaiac fecal occult blood tests in screening programs.
Mandel JS, Bond JH, Church TR, et al.: Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N.Engl.J.Med., 1993; 328: 1365–1371
Mandel JS, Church TR, Bond JH, et al.: The effect of fecal occult-blood screening on the incidence of colorectal cancer. N.Engl.J.Med., 2000; 343: 1603–1607
Atkin WS, Edwards R, Kralj-Hans I, et al.; UK Flexible Sigmoidoscopy Trial Investigators: Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet. 2010;375:1624–33. This is the first unbiased population-based randomized trial to show without doubt that polyp detection and removal reduces incidence and mortality from colorectal cancer.
Bossuyt PM, Irwig L, Craig J, Glasziou P: Comparative accuracy: assessing new tests against existing diagnostic pathways BMJ. 2006;332:1089–1092.
Pepe MS, Etzioni R, Feng S, et al.: Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061.
St John DJB, Young GP, Alexeyeff MA, et al.: Evaluation of new occult blood tests for detection of colorectal neoplasia. Gastroenterology 1993, 104, 1661–1668.
Smith A, Young GP, Cole SR, Bampton P: Comparison of a brush-sampling fecal immunochemical test for hemoglobin with a sensitive guaiac-based fecal occult blood test in detection of colorectal neoplasia. Cancer. 2006;107:2152–2159
••Young GP, Allison J: Colorectal cancer screening. In Textbook of Gastroenterology (5th edition), Eds Yamada T, Alpers D Kalloo AN, Kaplowitz N, Owyang C, Powell DW. Lippincott Williams and Wilkins, Philadelphia, 2009, 1598–1610. This is a comprehensive review of the evidence for screening for colorectal cancer and the pros and cons of the screening tests. This review takes a population-based approach with emphasis on engaging populations and feasibility.
Jones S, Chen WD, Parmigiani G, et al.: Comparative lesion sequencing provides insights into tumor evolution. Proc.Natl.Acad.Sci.U.S.A, 2008; 105: 4283–4288
••Levin B, Lieberman DA, McFarland B, et al.; American Cancer Society Colorectal Cancer Advisory Group; US Multi-Society Task Force; American College of Radiology Colon Cancer Committee: Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology 2008;134:1570–95. These are the US guidelines on screening, with emphasis on polyps as a key strategy.
Atkin WS, Morson BC, Cuzick J: Long-term risk of colorectal cancer after excision of rectosigmoid adenomas. New Engl J Med 1992 Mar 5;326(10):658–62.
Winawer SJ, Zauber AG, O’Brien MJ, et al.: Randomized comparison of surveillance intervals after colonoscopic removal of newly diagnosed adenomatous polyps. The National Polyp Study Workgroup. New Engl J Med 1993 Apr 1;328(13):901–6.
Strope SA, Andriole GL: Prostate cancer screening: current status and future perspectives. Nat Rev Urol 2010 Sep;7(9):487–93.
Hermsen M, Postma C, Baak J, et al.: Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology, 2002;123:1109–1119.
Carvalho B, Postma C, Mongera S, et al.: Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut, 2009 Jan;58(1):79–89
••Bosch LJW, Carvalho B, Fijneman RJA, et al.: Molecular tests for colorectal cancer screening. Clin Colorectal Cancer 2011 Jan; in press. This is the best and most up-to-date review of the detail of molecular tests for colorectal cancer screening, having a more scientific focus. It also includes information on blood tests for screening.
Osborn NK , Ahlquist DA: Stool screening for colorectal cancer: molecular approaches. Gastroenterology, 2005; 128: 192–206
Théry C, Ostrowski M, Segura E: Membrane vesicles as conveyors of immune response. Nat Rev Immunol. 2009;9:581–593.;
Ahlquist DA, Skoletsky JE, Boynton KA, et al.: Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel. Gastroenterology, 2000; 119: 1219–1227
Boynton KA, Summerhayes IC, Ahlquist DA, et al.: DNA integrity as a potential marker for stool-based detection of colorectal cancer. Clin.Chem., 2003; 49: 1058–1065
Schuebel KE, Chen W, Cope L, et al.: Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS.Genet., 2007; 3: 1709–1723
•Calistri D, Rengucci C, Molinari C, et al.: Quantitative fluorescence determination of long-fragment DNA in stool as a marker for the early detection of colorectal cancer. Cell Oncol., 2009; 31: 11–17. This provides convincing evidence for the value of assaying DNA integrity using improved methods.
Imperiale TF, Ransohoff DF, Itzkowitz SH, et al.: Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N.Engl.J.Med., 2004; 351: 2704–2714]
••Ahlquist DA, Sargent DJ, Loprinzi CL, et al.: Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann.Intern.Med., 2008; 149: 441–50. This is a valuable comparison of fecal DNA tests with guaiac-faecal occult blood tests.
•Itzkowitz S, Brand R, Jandorf L, et al.: A simplified, noninvasive stool DNA test for colorectal cancer detection. Am.J.Gastroenterol., 2008; 103: 2862–2870. This is an early paper showing the value of vimentin in detection but the problems with specificity as one seeks better sensitivity.
Chen WD, Han ZJ, Skoletsky J, et al.: Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J.Natl.Cancer Inst., 2005; 97: 1124–1132
Tagore KS, Lawson MJ, Yucaitis JA, et al.: Sensitivity and specificity of a stool DNA multitarget assay panel for the detection of advanced colorectal neoplasia. Clin.Colorectal Cancer, 2003; 3: 47–53
Slaby O, Svoboda M, Michalek J, Vyzula R: MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Molecular Cancer 2009;8:102
•LaPointe LC, Dunne R, Brown GS, et al.: Map of Differential Transcript Expression in the Normal Human Large Intestine. Physiological Genomics, 2008, 33:50–64. This is the only paper addressing gene expression patterns along the normal colon. Clearly, there is considerable variation in the normal state.
Kanaoka S, Yoshida K, Miura N, et al.: Potential usefulness of detecting cyclooxygenase 2 messenger RNA in feces for colorectal cancer screening. Gastroenterology, 2004; 127: 422–427
Leung WK, To KF, Man EP, et al.: Detection of hypermethylated DNA or cyclooxygenase-2 messenger RNA in fecal samples of patients with colorectal cancer or polyps. Am.J.Gastroenterol., 2007; 102: 1070–1076
Koga Y, Yasunaga M, Moriya Y, et al.: Detection of colorectal cancer cells from feces using quantitative real-time RT-PCR for colorectal cancer diagnosis. Cancer Sci., 2008; 99: 1977–1983
Takai T, Kanaoka S, Yoshida K, et al.: Fecal cyclooxygenase 2 plus matrix metalloproteinase 7 mRNA assays as a marker for colorectal cancer screening. Cancer Epidemiol.Biomarkers Prev., 2009; 18: 1888–1893
Yajima S, Ishii M, Matsushita H, et al.: Expression profiling of fecal colonocytes for RNA-based screening of colorectal cancer. Int.J.Oncol., 2007; 31: 1029–1037
•Mulder SA, van Leerdam ME, van Vuuren AJ, et al.: Tumor pyruvate kinase isoenzyme type M2 and immunochemical fecal occult blood test: performance in screening for colorectal cancer. Eur.J.Gastroenterol.Hepatol., 2007; 19: 878–882. An example of a useful stool-based protein marker but, as with haptoglobin (see [42] below), without obvious major advantage over stool hemoglobin.
Shastri YM, Povse N, Schroder O, et al.: Comparison of a novel fecal marker--fecal tumor pyruvate kinase type M2 (M2-PK) with fecal calprotectin in patients with inflammatory bowel disease: a prospective study. Clin.Lab, 2008; 54: 389–390
Haug U, Hundt S, Brenner H: Sensitivity and specificity of faecal tumour M2 pyruvate kinase for detection of colorectal adenomas in a large screening study. Br.J.Cancer, 2008; 99: 133135
Karl J, Wild N, Tacke M, et al.: Improved diagnosis of colorectal cancer using a combination of fecal occult blood and novel fecal protein markers. Clin.Gastroenterol.Hepatol., 2008; 6: 1122–1128
Xing PX, Young GP, Ho D, et al.: A new approach to fecal occult blood testing based on the detection of haptoglobin. Cancer, 1996; 78: 48–56
Sieg A, Thoms C, Luthgens K, et al.: Detection of colorectal neoplasms by the highly sensitive hemoglobin-haptoglobin complex in feces. Int.J.Colorectal Dis., 1999; 14: 267–271
von Roon AC, Karamountzos L, Purkayastha S, et al.: Diagnostic precision of fecal calprotectin for inflammatory bowel disease and colorectal malignancy. Am.J.Gastroenterol., 2007; 102: 803–813
Disclosure
No potential conflicts of interest relevant to this article were reported.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Young, G.P., Bosch, L.J.W. Fecal Tests: From Blood to Molecular Markers. Curr Colorectal Cancer Rep 7, 62–70 (2011). https://doi.org/10.1007/s11888-010-0084-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11888-010-0084-8