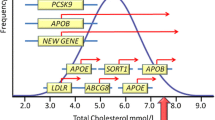
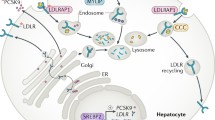
Abstract
Familial hypercholesterolemia (FH) is a genetic disorder characterized by elevated low-density lipoprotein (LDL) cholesterol and premature cardiovascular disease, with a prevalence of approximately 1 in 200–500 for heterozygotes in North America and Europe. Monogenic FH is largely attributed to mutations in the LDLR, APOB, and PCSK9 genes. Differential diagnosis is critical to distinguish FH from conditions with phenotypically similar presentations to ensure appropriate therapeutic management and genetic counseling. Accurate diagnosis requires careful phenotyping based on clinical and biochemical presentation, validated by genetic testing. Recent investigations to discover additional genetic loci associated with extreme hypercholesterolemia using known FH families and population studies have met with limited success. Here, we provide a brief overview of the genetic determinants, differential diagnosis, genetic testing, and counseling of FH genetics.

Similar content being viewed by others
Notes
Note: For the purposes of this article, it can be assumed that “FH” refers to the heterozygous state as it is the more common form of FH; homozygous FH (HoFH) will be specified in the text.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Fahed A, Safa R, Haddad F, et al. Homozygous familial hypercholesterolemia in Lebanon: a genotype/phenotype correlation. Mol Genet Metab. 2011;102:181–8.
Goldstein J, Schrott H, Hazzard W, et al. Hyperlipidemia in coronary heart disease: genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J Clin Invest. 1973;52:1544–68.
Rader D, Cohen J, Hobbs H. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111(12):1795–803.
Austin A, Hutter C, Zimmern R, Humphries S. Genetic causes of monogenic heterozygous familial hypercholesterolemia: a HuGE prevalence review. Am J Epidemiol. 2004;160:407–20.
Seftel H, Baker S, Jenkins T, Mendelsohn D. Prevalence of familial hypercholesterolemia in Johannesburg Jews. Am J Med Genet. 1989;34:545–7.
Nordestgaard B, Chapman M, Humphries S, et al. Familial hypercholesterolemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease. Eur Heart J. 2013;34(45):3478–90. This is the Consensus Statement of the European atherosclerosis Society discussing the underdiagnoses of FH, its genetics, and screening approach.
Seidah NG, Awan Z, Chrétien M, Mbikay M. PCSK9: a key modulator of cardiovascular health. Circ Res. 2014;114:1022–36. A thorough review of the biological and genetic characteristics of PCSK9 including animal models and the possible clinical utility.
Cuchel M, Bruckert E, Ginsberg H, et al. Homozygous familial hypercholesterolaemia: new insights and guidance for clinicians to improve detection and clinical management. A position paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Hypercholesterolaemia of the European Atherosclerosis Society. Eur Heart J. 2014;35:2146–57. This is the Consensus Statement of the European atherosclerosis Society and provides an updated approach to genetics and screening of familial hypercholesterolemia.
Tada H, Kawashiri M, Ohtani R, et al. A novel type of familial hypercholesterolemia: double heterozygous mutations in LDL receptor and LDL receptor adaptor protein 1 gene. Atherosclerosis. 2011;219:663–6.
Marduel M, Carrie A, Sassolas A, et al. Molecular spectrum of autosomal dominant hypercholesterolemia in France. Hum Mutat. 2010;31:E1811–1824.
Ejarque I, Real J, Martinez-Hervas S, et al. Evaluation of clinical diagnosis criteria of familial ligand defective apoB 100 and lipoprotein phenotype comparison between LDL receptor gene mutations affecting ligand-binding domain and the R3500Q mutation of the apoB gene in patients from a South European population. Transl Res. 2008;151:162–7.
Benlian P, de Gennes J, Dairou F, et al. Phenotypic expression in double heterozygotes for familial hypercholesterolemia and familial defective apolipoprotein B-100. Hum Mutat. 1996;7:340–5.
Taylor A, Bayly G, Patel K, et al. A double heterozygote for familial hypercholesterolaemia and familial defective apolipoprotein B-100. Ann Clin Biochem. 2010;47:487–90.
Pisciotta L, Oliva P, Cefalu A, et al. Additive effect of mutations in LDLR and PCSK9 genes on the phenotype of familial hypercholesterolemia. Atherosclerosis. 2006;186:433–40.
Noguchi T, Katsuda S, Kawashiri M, et al. The E32K variant of PCSK9 exacerbates the phenotype of familial hypercholesterolaemia by increasing PCSK9 function and concentration in the circulation. Atherosclerosis. 2010;210:166–72.
Soufi M, Rust S, Walter M, Schaefer J. A combined LDL receptor/LDL receptor adaptor protein 1 mutation as the cause for severe familial hypercholesterolemia. Gene. 2013;521:200–3.
Garcia-Garcia A, Ivorra C, Martinez-Hervas S, et al. Reduced penetrance of autosomal dominant hypercholesterolemia in a high percentage of families: importance of genetic testing in the entire family. Atherosclerosis. 2011;218:423–30.
Ahmad Z, Adams-Huet B, Chen C, Garg A. Low prevalence of mutations in known loci for autosomal dominant hypercholesterolemia in multi-ethnic patient cohort. Circ Cardiovasc Genet. 2012;5:666–75.
Leigh S, Foster A, Whittall R, Hubbart C, Humphries S. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann Hum Genet. 2008;72:485–98.
Alonso R, Mata N, Castillo S, et al. Cardiovascular disease in familial hypercholesterolaemia: influence of low-density lipoprotein receptor mutation type and classic risk factors. Atherosclerosis. 2008;200:315–21.
Hopkins P, Toth P, Ballantyne C, Rader D. Familial hypercholesterolemias: prevalence, genetics, diagnosis and screening recommendations from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J Clin Lipidol. 2011;5:S9–17.
Soutar A, Naoumova R. Mechanisms of disease: genetic causes of familial hypercholesterolemia. Nat Clin Pract Cardiovasc Med. 2007;4:214–25.
Usifo E, Leigh S, Whittall R, et al. Low-density lipoprotein receptor gene familial hypercholesterolemia variant database: update and pathological assessment. Ann Hum Genet. 2012;76:387–401.
Gudnason V, Day I, Humphries S. Effect on plasma lipid levels of different classes of mutation in the low-density lipoprotein receptor gene in patients with familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 1994;14:1717–22.
Hobbs H, Brown M, Goldstein J. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–66.
Pisciotta L, Cantafora A, De Stefano F, Langheim S, Calandra S, Bertolini S. A “de novo” mutation of the LDL-receptor gene as the cause of familial hypercholesterolemia. Biochim Biophys Acta. 2002;1587:7–11.
Hobbs H, Russell D, Brown M, et al. The LDL receptor locus and familial hypercholesterolemia: mutational analysis of a membrane protein. Annu Rev Genet. 1990;24:133–70.
De Castro-Oros I, Pampin S, Bolado-Carrancio A, et al. Functional analysis of LDLR promoter and 5’ UTR mutations in subjects with clinical diagnosis of familial hypercholesterolemia. Hum Mutat. 2011;32:868–72.
Guardamagna O, Restagno G, Rolfo E, et al. The type of LDLR gene mutation predicts cardiovascular risk in children with familial hypercholesterolemia. J Pediatr. 2009;155:199–204.
Ten Kate G, Neefjes L, Dedic A, et al. The effect of LDLR-negative genotype on CT coronary atherosclerosis in asymptomatic statin treated patients with heterozygous familial hypercholesterolemia. Atherosclerosis. 2013;227:334–41.
Whitfield A, Barrett H, van Bockxmeer F, Burnett J. Lipid disorder and mutations in the APOB gene. Clin Chem. 2004;50:1725–32.
Blackhart B, Ludwig E, Pierotti V, et al. Structure of the human apolipoprotein B gene. J Biol Chem. 1986;261:15364–7.
Myant N. Familial defective apolipoprotein B-100: a review, including some comparisons with familial hypercholesterolaemia. Atherosclerosis. 1993;104:1–18.
Boren J, Ekstrom U, Agren B, Nilsson-Ehle P, Innerarity T. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J Biol Chem. 2001;276:9214–8.
Innerarity T, Mahley R, Weisgraber K, et al. Familial defective apolipoprotein B-100: a mutation of apolipoprotein B that causes hypercholesterolemia. J Lipid Res. 1990;31:1337–49.
Pullinger C, Hennessy L, Chatterton J, et al. Familial ligand-defective apolipoprotein B—identification of a new mutation that decreases LDL receptor binding affinity. J Clin Invest. 1995;95:1225–34.
Soufi M, Sattler A, Maerz W, et al. A new but frequent mutation of APOB-100APOB His 3543Tyr. Atherosclerosis. 2004;174:11–6.
Tai D, Pan J, Lee-Chen G. Identification and haplotype analysis of apolipoprotein B-100 Arg3500 → Trp mutation in hyperlipidemic Chinese. Clin Chem. 1998;44:1659–65.
Al-Khateeb A, Mohd M, Yusof Z, Zilfalil B. Molecular description of familial defective APOB-100 in Malaysia. Biochem Genet. 2013;51:811–23.
Motazacker MM, Pirruccello J, Huijgen R, et al. Advances in genetics show the need for extending screening strategies for autosomal dominant hypercholesterolaemia. Eur Heart J. 2012;33:1360–6.
Thomas ER, Atanur SS, Norsworthy PJ, et al. Identification and biochemical analysis of a novel APOB mutation that causes autosomal dominant hypercholesterolemia. Mol Genet Genomic Med. 2013;1:155–61.
Cohen J, Boerwinkle E, Mosley Jr T, Hobbs H. Sequence variations in PCSK9, low LDL and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72.
Abifadel M, Elbitar S, El Khoury P, et al. Living the PCSK9 adventure: from the identification of a new gene in familial hypercholesterolemia towards a potential new class of anticholesterol drugs. Curr Atheroscler Rep. 2014;16:439.
Cariou B, Le May C, Costet P. Clinical aspects of PCSK9. Atherosclerosis. 2011;216:258–65.
Pandit S, Wisniewski D, Santoro J, et al. Functional analysis of sites within PCSK9 responsible for hypercholesterolemia. J Lipid Res. 2008;49:1333–43.
De Castro-Oros I, Pocovi M, Civeira F. The genetic basis of familial hypercholesterolemia: inheritance, linkage and mutations. Appl Clin Genet. 2010;3:52–64.
Mabuchi H, Nohara A, Noguchi T, et al. Genotypic and phenotypic features in homozygous familial hypercholesterolemia caused by proprotein convertase subtilisin/kexin type 9 (PCSK9) gain-of-function mutation. Atherosclerosis. 2014;236:54–61.
Pisciotta L, Oliva C, Pes G, et al. Autosomal recessive hypercholesterolemia (ARH) and homozygous familial hypercholesterolemia (FH): a phenotypic comparison. Atherosclerosis. 2006;188:398–405.
Garcia K, Wilund K, Arca M, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–8.
Rahalkar A, Hegele R. Monogenic pediatric dyslipidemias: classification, genetics and clinical spectrum. Mol Genet Metab. 2008;93:282–94.
Filigheddu F, Quagliarini F, Campagna F, et al. Prevalence and clinical features of heterozygous carriers of autosomal recessive hypercholesterolemia in Sardinia. Atherosclerosis. 2009;207:162–7.
Naoumova R, Neuwirth C, Lee P, et al. Autosomal recessive hypercholesterolaemia: long-term follow up and response to treatment. Atherosclerosis. 2004;174:165–72.
Awan Z, Choi HY, Stitziel N, et al. APOE p.Leu167del mutation in familial hypercholesterolemia. Atherosclerosis. 2013;231:218–22.
Marduel M, Ouguerram K, Serre V, et al. Description of a large family with autosomal dominant hypercholesterolemia associated with the APOE p.Leu167del mutation. Hum Mutat. 2013;34:83–7.
Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through IDOL-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–4.
Fahed A, Nemer G. Familial hypercholesterolemia: the lipids or the genes? Nutr Metab (Lond). 2011;8:1–12.
Zhang L, Reue K, Fong L, Young S, Tontonoz P. Feedback regulation of cholesterol uptake by the LDL-IDOL-LDLR axis. Arterioscler Thromb Vasc Biol. 2012;32:2541–6.
Weissglas-Volkov D, Calkin A, Tusie-Luna T, et al. The N342 MYLIP polymorphism is associated with high total cholesterol and increased LDL receptor degradation in humans. J Clin Invest. 2011;121:3062–71.
Marques-Pinheiro A, Marduel M, Rabes JP, et al. A fourth locus for autosomal dominant hypercholesterolemia maps at 16q22.1. Eur J Hum Genet. 2010;18:1236–42.
Fouchier S, Dallinga-Thie G, Meijers J, et al. Mutations in STAP1 are associated with autosomal dominant hypercholesterolemia. Circ Res. 2014;115:552–5.
Futema M, Plagnol V, Li K, et al. Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J Med Genet. 2014;51:537–44.
Lange L, Hu Y, Zhang H, et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94:233–45.
Bjorkhem I. Cerebrotendinous xanthomatosis. Curr Opin Lipidol. 2013;24:283–7.
Mignarri A, Gallus GN, Dotti MT, Federico A. A suspicion index for early diagnosis and treatment of cerebrotendinous xanthomatosis. J Inherit Metab Dis. 2014;37:421–9.
Inanloorahatloo K, Zand Parsa AF, Huse K, Rasooli P, Davaran S, Platzer M, et al. Mutation in CYP27A1 identified in family with coronary artery disease. Eur J Med Genet. 2013;56:655–60.
Khan AO, Aldahmesh MA, Mohamed JY, Alkuraya FS. Juvenile cataract morphology in 3 siblings not yet diagnosed with cerebrotendinous xanthomatosis. Ophthalmology. 2013;120:956–60.
Monson DM, DeBarber AE, Bock CJ, Anadiotis G, Merkens LS, Steiner RD, et al. Cerebrotendinous xanthomatosis: a treatable disease with juvenile cataracts as a presenting sign. Arch Ophthalmol. 2011;129:1087–8.
Martini G, Mignarri A, Ruvio M, Valenti R, Franci B, Del Puppo M, et al. Long-term bone density evaluation in cerebrotendinous xanthomatosis: evidence of improvement after chenodeoxycholic acid treatment. Calcif Tissue Int. 2013;92:282–6.
Fraidakis MJ. Psychiatric manifestations in cerebrotendinous xanthomatosis. Transl Psychiatry. 2013;3:e302.
Lorincz MT, Rainier S, Thomas D, Fink JK. Cerebrotendinous xanthomatosis: possible higher prevalence that previously recognized. Arch Neurol. 2005;62:1459–63.
Reshef A, Meiner V, Berginer VM, Leitersdorf E. Molecular genetics of cerebrotendinous xanthomatosis in Jews of north African origin. J Lipid Res. 1994;35:478–83.
Leitersdorf E, Safadi R, Meiner V, Reshef A, Björkhem I, Friedlander Y, et al. Cerebrotendinous xanthomatosis in the Israeli Druze: molecular genetics and phenotypic characteristics. Am J Hum Genet. 1994;55:907–15.
Batta AK, Salen G, Shefer S, Tint GS, Batta M. Increased plasma bile alcohol glucuronides in patients with cerebrotendinous xanthomatosis: effect of chenodeoxycholic acid. J Lipid Res. 1987;28:1006–12.
DeBarber AE, Connor WE, Pappu AS, Merkens LS, Steiner RD. ESI-MS/MS quantification of 7alpha-hydroxy-4-cholesten-3-one facilitates rapid, convenient diagnostic testing for cerebrotendinous xanthomatosis. Clin Chim Acta. 2010;411:43–8.
Matysik S, Orsó E, Black A, Ahrens N, Schmitz G. Monitoring of 7a-hydroxy-4-cholesten-3-one during therapy of cerebrotendinous xanthomatosis: a case report. Chem Phys Lipids. 2011;164:530–4.
DeBarber AE, Luo J, Star-Weinstock M, Purkayastha S, Geraghty MT, Chiang JP, et al. A blood test for cerebrotendinous xanthomatosis with potential for disease detection in newborns. J Lipid Res. 2014;55:146–54.
Diekstra FP, Saris CG, van Rheenen W, Franke L, Jansen RC, van Es MA, et al. Mapping of gene expression reveals CYP27A1 as a susceptibility gene for sporadic ALS. PLoS One. 2012;7:e35333.
Berginer VM, Gross B, Morad K, Kfir N, Morkos S, Aaref S, et al. Chronic diarrhea and juvenile cataracts: think cerebrotendinous xanthomatosis and treat. Pediatrics. 2009;123:143–7.
Bhattacharyya AK, Connor WE. Beta-sitosteroemia and xanthomatosis: a newly described lipid storage disease in two sisters. J Clin Invest. 1974;53:1033–43.
Lu K, M-h L, Hazard S, Brooks-Wilson A, Hidaka H, Kojima H, et al. Two genes that map to the STSL locus cause sitosterolemia: genomic structure and spectrum of mutations involving sterolin-1 and sterolin-2, encoded by ABCG5 and ABCG8, respectively. Am J Hum Genet. 2001;69:278–90.
Yu XH, Qian K, Jiang N, Zheng XL, Cayabyab FS, Tang CK. ABCG5/ABCG8 in cholesterol excretion and atherosclerosis. Clin Chim Acta. 2014;428:82–8.
Escola-Gil JC, Quesada H, Julve J, Martin-Campos JM, Cedo L, Blanco-Vaca F. Sitosterolemia: diagnosis, investigation and management. Curr Atheroscler Rep. 2014;16:424.
Lin DS, Steiner RD, Merkens LS, Pappu AS, Coner WE. The effects of sterol structure upon sterol esterification. Atherosclerosis. 2010;208:155–60.
Patel SB. Recent advances in understanding the STSL locus and ABCG5/ABCG8 biology. Curr Opin Lipidol. 2014;25:169–75.
Lee MH, Lu K, Patel SB. Genetic basis of sitosterolemia. Curr Opin Lipidol. 2001;12:141–9.
Othman RA, Myrie SB, Jones PJ. Non-cholesterol sterols and cholesterol metabolism in sitosterolemia. Atherosclerosis. 2013;231:291–9.
Hansel B, Carrié A, Brun-Druc N, Leclert G, Chantepie S, Coiffard AS, et al. Premature atherosclerosis is not systematic in phytosterolemic patients: severe hypercholesterolemia as a confounding factor in five subjects. Atherosclerosis. 2014;234:162–8.
Silbernagel G, Chapman MJ, Genser B, Kleber ME, Fauler G, Scharnagl H, et al. High intestinal cholesterol absorption is associated with cardiovascular disease and risk alleles in ABCG8, and ABO. J Am Coll Cardiol. 2013;62:291–9.
Teupser D, Baber R, Scholz M, Illig T, Gieger C, Holdt LM, et al. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ Cardiovasc Genet. 2010;3:331–9.
Wang Z, Cao L, Su Y, Wang G, Wang R, Yu Z, et al. Specific macrothrombocytopenia/hemolytic anemia associated with sitosterolemia. Am J Hematol. 2014;89:320–4.
Salen G, von Bergmann K, Lutjohann D, Kwiterovich P, Kane J, Patel SB, et al. Ezetimibe effectively reduces plasma plant sterols in patients with sitosterolemia. Circulation. 2004;109:966–71.
Tang W, Ma Y, Ioannou YA, Davies YP, Yu L. Genetic inactivation of NPLC1L1 protects against sitosterolemia in mice lacking ABCG5/ABCG8. J Lipid Res. 2009;50:293–300.
Niu DM, Chong KW, Hsu JH, Wu TJ, Huang CH, Lo MY, et al. Clinical observations, molecular genetic analysis and treatment of sitosterolemia in infants and children. J Inherit Metab Dis. 2010;33:437–43.
Lutjohann D, von Bergmann K, Sirah W, MacDonell G, Johnson-Levonas AO, Shah A, et al. Long term efficacy and safety of ezetimibe 10 mg in patients with homozygous sitosterolemia: a 2-year, open label extension trial. Int J Clin Prac. 2008;62:1499–510.
Myrie SB, Mymin D, Triggs-Raine B, Jones PJ. Serum lipid, plant sterols, and cholesterol kinetic responses to plant sterol supplementation in phytosterolemia heterozygotes and control individuals. Am J Clin Nutr. 2012;95:837–44.
Kanaji T, Kanaji S, Montgomery RR, Patel SB, Newman PJ. Platelet hyperreactivity explains the bleeding abnormality and macrothrombocytopenia in a murine model of sitosterolemia. Blood. 2013;122:2732–42.
McDaniel AL, Alger HM, Sawyer JK, Kelley KL, Kock ND, Brown JM, et al. Phytosterol feeding causes toxicity in ABCG5/G8 knockout mice. Am J Pathol. 2013;182:1131–8.
Aslanidis C, Klima H, Lackner KJ, Schmitz G. Genomic organization of the human lysosomal acid lipase gene (LIPA). Genomics. 1994;20:329–31.
Fouchier SW, Defesche JC. Lysosomal acid lipase A and the hypercholesterolaemic phenotype. Curr Opin Lipidol. 2013;24:332–8.
Bernstein DL, Hulkova H, Bialer MG, Desnick RJ. Cholesterol ester storage disease: review of the findings in 135 reported patients with an underdiagnosed disease. J Hepatol. 2013;58:1230–43.
Elleder M, Chlumska A, Hyanek J, et al. Subclinical course of cholesteryl ester storage disease in an adult with hypercholesterolemia, accelerated atherosclerosis and liver cancer. J Hepatol. 2000;32:528–34.
Stitziel NO, Fouchier SW, Sjouke B, Peloso GM, Moscoso AM, Auer PL, et al. National Heart, Lung, and Blood Institute GO Exome Sequencing Project. Arterioscler Thromb Vasc Biol. 2013;33:2909–14.
Reiner Z, Guardamagna O, Nair D, Soran H, Hovingh K, Bertolini S, et al. Lysosomal acid lipase deficiency-an under-recognized cause of dyslipidaemia and liver dysfunction. Atherosclerosis. 2014;235:21–30.
Scott SA, Liu B, Nazarenko I, Martis S, Kozkitina J, Yang Y, et al. Frequency of the cholesterol ester storage disease common LIPA E8SJM mutation (c.894G>A) in various racial and ethnic groups. Hepatology. 2013;58:958–65.
Reynolds T. Cholesterol ester storage disease: a rare and possibly treatable cause of premature vascular disease and cirrhosis. J Clin Pathol. 2013;66:818–923.
Hamilton J, Jones I, Srivastava R, Galloway P. A new method for the measurement of lysosomal acid lipase in dried blood spots using the inhibitor lalistat 2. Clin Chim Acta. 2012;413:1207–10.
Balwani M, Breen C, Enns GM, Deegan PB, Honzík T, Jones S, et al. Clinical effect and safety profile of recombinant human lysosomal acid lipase in patients with cholesteryl ester storage disease. Hepatology. 2013;58:950–7.
National Society of Genetic Counselors' Definition Task Force, Resta R, Biesecker BB, Bennett RL, Blum S, Hahn SE, et al. A new definition of genetic counseling: National Society of Genetic Counselors’ Task Force Report. J Genet Couns. 2006;15(2):77–83.
Sturm AC. The role of genetic counselors for patients with familial hypercholesterolemia. Curr Genet Med Rep. 2014;2:68–74.
ACMG Board of Directors. Points to consider in the clinical application of genomic sequencing. Genet Med. 2012;14(8):759–61.
Miller C, Krautscheid P, Baldwin E, et al. Genetic counselor review of genetic test orders in a reference laboratory reduces unnecessary testing. Am J Med Genet. 2014;164A:1094–101.
Leighton J, Valverde K, Berhardt B. The general public’s understanding and perception of direct-to-consumer genetic test results. Public Health Genomics. 2012;15:11–21.
Hilgart J, Mercer J, Thirlaway K. Individuals’ experiences of, and responses to, a negative genetic test result for familial hypercholesterolemia. J Health Psychol. 2013;18:339–49.
Santos RD, Maranhao RC. What is new in familial hypercholestgerolemia? Curr Opin Lipidol. 2014;25:183–8.
Marks D, Wonderling D, Thorogood M, et al. Cost effectiveness analysis of different approaches of screening for familial hypercholesterolemia. BMJ. 2002;324:1303.
Leren T, Finborud T, Manshaus T, Ose L, Berge K. Diagnosis of familial hypercholesterolemia in general practice using clinical diagnostic criteria or genetic testing as part of cascade genetic screening. Community Genet. 2008;11:26–35.
Watts GF, Gidding S, Wierzbicki AS, Toth PP, Alonso R, Brown WV, et al. Integrated guidance on the care of familial hypercholesterolaemia from the International FH Foundation. Int J Cardiol. 2014;171:309–25.
Van der Roest W, Pennings J, Bakker M, van den Berg M, van Tintelen J. Family letters are an effective way to inform relatives about inherited cardiac disease. Am J Med Genet A. 2009;149A:357–63.
Slimani A, Jelassi A, Jguirim I, Najah M, Rebhi L, Omezzine A, et al. Effect of mutations in LDLR and PCSK9 genes on phenotypic variability in Tunisian familial hypercholesterolemia patients. Atherosclerosis. 2012;222(1):158–66.
Mabuchi H, Nohara A, Noguchi T, et al. Molecular genetic epidemiology of homozygous familial hypercholesterolemia in the Hokuriku district of Japan. Atherosclerosis. 2011;214:404–7.
Tichý L, Freiberger T, Zapletalová P, Soška V, Ravčuková B, Fajkusová L. The molecular basis of familial hypercholesterolemia in the Czech Republic: spectrum of LDLR mutations and genotype-phenotype correlations. Atherosclerosis. 2012;223(2):401–8.
Diakou M, Miltiadous G, Xenophontos SL, Manoli P, Cariolou MA, Elisaf M. Spectrum of LDLR gene mutations, including a novel mutation causing familial hypercholesterolaemia, in North-western Greece. Eur J Intern Med. 2011;22(5):e55–9.
Bertolini S, Pisciotta L, Rabacchi C, Cefalù AB, Noto D, Fasano T, et al. Spectrum of mutations and phenotypic expression in patients with autosomal dominant hypercholesterolemia identified in Italy. Atherosclerosis. 2013;227(2):342–8.
Medeiros AM, Alves AC, Francisco V, Bourbon M, Investigators of the Portuguese FH Study. Update of the Portuguese Familial Hypercholesterolaemia Study. Atherosclerosis. 2010;21(2):553–8.
Palacios L, Grandoso L, Cuevas N, et al. Molecular characterization of familial hypercholesterolemia in Spain. Atherosclerosis. 2012;221:137–42.
Acknowledgements
We would like to acknowledge Ashley Kuharchik (CPhT) for the use of her graphic design skills. We would also like to thank Ingrid Glurich, PhD and Marie Fleisner for their editorial assistance in preparing this manuscript.
Compliance with Ethics Guidelines
ᅟ
Conflict of Interest
Ariel Brautbar, Emili Leary, Kristen Rasmussen, Don P. Wilson, Robert D. Steiner, and Salim Virani declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on New Drugs Approved for Homozygous FH
Rights and permissions
About this article
Cite this article
Brautbar, A., Leary, E., Rasmussen, K. et al. Genetics of Familial Hypercholesterolemia. Curr Atheroscler Rep 17, 20 (2015). https://doi.org/10.1007/s11883-015-0491-z
Published:
DOI: https://doi.org/10.1007/s11883-015-0491-z
Keywords
- Familial hypercholesterolemia (FH)
- Low-density lipoprotein receptor (LDLR)
- Apolipoprotein B (APOB)
- Familial defective apolipoprotein B-100 (FDB)
- Proprotein convertase subtilisin/kexin type 9 (PCSK9)
- Autosomal recessive hypercholesterolemia (ARH)
- LDL receptor adapter protein 1 (LDLRAP1)
- Genetic counseling (GC)
- Cascade testing
- Penetrance
- Cerebrotendinous xanthomatosis (CTX)
- Sitosterolemia
- Cholesterol ester storage disease (CESD)