Abstract
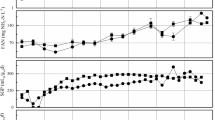
This study assessed the effects of reducing hydraulic retention times (HRTs) from 21 days to 10.5 days when anaerobically co-digesting pig manure and food waste. Continuously stirred tank reactors of 3.75 L working volume were operated in triplicate at 42°C. Digester HRT was progressively decreased from 21 to 15 days to 10.5 days, with an associated increase in organic loading rate (OLR) from 3.1 kg volatile solids (VS)·m–3·day–1 to 5.1 kg VS·m–3·day–1 to 7.25 kg VS·m–3·day–1. Reducing HRT from 21 days to 15 days caused a decrease in specific methane yields and VS removal rates. Operation at a HRT of 10.5 days initially resulted in the accumulation of isobutyric acid in each reactor. High throughput 16S rRNA gene sequencing revealed that this increase coincided with a shift in acidogenic bacterial populations, which most likely resulted in the increased isobutyric acid concentrations. This may in turn have caused the increase in relative abundance of Clocamonaceae bacteria, which syntrophically degrade non-acetate volatile fatty acids (VFAs) into H2 and CO2. This, along with the increase in abundance of other syntrophic VFA oxidizers, such as Spiorchatetes, suggests that VFA oxidation plays a role in digester operation at low HRTs. Reducing the HRT to below 21 days compromised the ability of the anaerobic digestion system to reduce enteric indicator organism counts below regulatory limits.

Similar content being viewed by others
References
Xie S, Wu G, Lawlor P G, Frost J P, Zhan X. Methane production from anaerobic co-digestion of the separated solid fraction of pig manure with dried grass silage. Bioresource Technology, 2012, 104: 289–297
Sundberg C, Al-Soud W A, Larsson M, Alm E, Yekta S S, Svensson B H, Sørensen S J, Karlsson A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiology Ecology, 2013, 85(3): 612–626
Dennehy C, Lawlor P G, Croize T, Jiang Y, Morrison L, Gardiner G E, Zhan X. Synergism and effect of high initial volatile fatty acid concentrations during food waste and pig manure anaerobic codigestion. Waste Management (New York, N.Y.), 2016, 56: 173–180
Batstone D, Tait S, Starrenburg D. Estimation of hydrolysis parameters in full-scale anerobic digesters. Biotechnology and Bioengineering, 2009, 102(5): 1513–1520
Stolze Y, Zakrzewski M, Maus I, Eikmeyer F, Jaenicke S, Rottmann N, Siebner C, Puhler A, Schluter A. Comparative metagenomics of biogas-producing microbial communities from production-scale biogas plants operating under wet or dry fermentation conditions. Biotechnology for Biofuels, 2015, 8(1): 14–20
Alsouleman K, Linke B, Klang J, Klocke M, Krakat N, Theuerl S. Reorganisation of a mesophilic biogas microbiome as response to a stepwise increase of ammonium nitrogen induced by poultry manure supply. Bioresource Technology, 2016, 208: 200–204
Lebuhn M, Hanreich A, Klocke M, Schlüter A, Bauer C, Pérez C M. Towards molecular biomarkers for biogas production from lignocellulose-rich substrates. Anaerobe, 2014, 29: 10–21
De Vrieze J, Saunders A M, He Y, Fang J, Nielsen P H, Verstraete W, Boon N. Ammonia and temperature determine potential clustering in the anaerobic digestion microbiome. Water Research, 2015, 75: 312–323
De Vrieze J, Raport L, Roume H, Vilchez-Vargas R, Jáuregui R, Pieper D H, Boon N. The full-scale anaerobic digestion microbiome is represented by specific marker populations. Water Research, 2016, 104: 101–110
McCutcheon G. A study of the dry matter and nutrient content of pig slurry. Dissertation for Master Degree. Dublin, Ireland: University College Dublin, 1997
Browne J D, Allen E, Murphy J D. Assessing the variability in biomethane production from the organic fraction of municipal solid waste in batch and continuous operation. Applied Energy, 2014, 128: 307–314
Anthonisen A, Loehr R, Prakasam T, Srinath E. Inhibition of nitrification by ammonia and nitrous acid. Journal-Water Pollution Control Federation, 1976, 48(5): 835–852
APHA. Standard Methods for the Examination of Water and Wastewater. Washington D. C.: APHA-AWWA-WEF, 1998
Batstone D J, Rodríguez J. Modelling Anaerobic Digestion Processes. In; Fang H P, Zhang T, eds. Anaerobic Biotechnology: Environmental Protection and Resource Recovery. London: Imperial College Press, 2015, 133–160
International Organization of Standardization. Microbiology of Food and Animal Feeding Stuffs-Horizontal Method for the Detection of Salmonella spp. Amendment 1: Annex D: Detection of Salmonella spp. in Animal Faeces and in Environmental Samples from the Primary Production Stage. Geneva, Switzerland: International Organization for Standardization, 2007
Caporaso J G, Lauber C L, Walters W A, Berg-Lyons D, Huntley J, Fierer N, Owens S M, Betley J, Fraser L, Bauer M, Gormley N, Gilbert J A, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal, 2012, 6(8): 1621–1624
De Santis T Z, Hugenholtz P, Larsen N, Rojas M, Brodie E L, Keller K, Huber T, Dalevi D, Hu P, Andersen G L. Greengenes, a chimerachecked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 2006, 72(7): 5069–5072
Fouhy F, Guinane C M, Hussey S, Wall R, Ryan C A, Dempsey E M, Murphy B, Ross R P, Fitzgerald G F, Stanton C, Cotter P D. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrobial Agents and Chemotherapy, 2012, 56(11): 5811–5820
Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience, 2013, 2(1): 16–22 doi:10.1186/2047-217X-2-16
Noike T, Endo G, Chang J E, Yaguchi J I, Matsumoto J I. Characteristics of carbohydrate degradation and the rate-limiting step in anaerobic digestion. Biotechnology and Bioengineering, 1985, 27(10): 1482–1489
Franke-Whittle I H, Walter A, Ebner C, Insam H. Investigation into the effect of high concentrations of volatile fatty acids in anaerobic digestion on methanogenic communities. Waste Management (New York, N.Y.), 2014, 34(11): 2080–2089
Maspolim Y, Zhou Y, Guo C, Xiao K, Ng W J. Comparison of single-stage and two-phase anaerobic sludge digestion systems–Performance and microbial community dynamics. Chemosphere, 2015, 140: 54–62
De Vrieze J, Gildemyn S, Vilchez-Vargas R, Jauregui R, Pieper D H, Verstraete W, Boon N. Inoculum selection is crucial to ensure operational stability in anaerobic digestion. Applied Microbiology and Biotechnology, 2015, 99(1): 189–199
Yamada T, Sekiguchi Y. Cultivation of uncultured Chloroflexi subphyla: significance and ecophysiology of formerly uncultured Chloroflexi “Subphylum I” with natural and biotechnological relevance. Microbes and Environments, 2009, 24(3): 205–216
Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Li T, Camacho P, Sghir A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME Journal, 2009, 3(6): 700–714
Ganidi N, Tyrrel S, Cartmell E. Anaerobic digestion foaming causes–A review. Bioresource Technology, 2009, 100(23): 5546–5554
Holdeman L V, Moore W E C. New genus, coprococcus, twelve new species, and emended descriptions of four previously described species of bacteria from human feces. International Journal of Systematic and Evolutionary Microbiology, 1974, 24(2): 260–277
Qiu Y L, Kuang X Z, Shi X S, Yuan X Z, Guo R B. Paludibacter jiangxiensis sp. nov., a strictly anaerobic, propionate-producing bacterium isolated from rice paddy field. Archives of Microbiology, 2014, 196(3): 149–155
Wu W M, Jain M K, Hickey R F, Zeikus J G. Perturbation of syntrophic isobutyrate and butyrate degradation with formate and hydrogen. Biotechnology and Bioengineering, 1996, 52(3): 404–411
Wu W M, Jain M K, Zeikus J G. Anaerobic degradation of normaland branched-chain fatty acids with four or more carbons to methane by a syntrophic methanogenic triculture. Applied and Environmental Microbiology, 1994, 60(7): 2220–2226
Hagen L H, Vivekanand V, Linjordet R, Pope P B, Eijsink V G H, Horn S J. Microbial community structure and dynamics during codigestion of whey permeate and cow manure in continuous stirred tank reactor systems. Bioresource Technology, 2014, 171: 350–359
Lee S H, Park J H, Kang H J, Lee Y H, Lee T J, Park H D. Distribution and abundance of Spirochaetes in full-scale anaerobic digesters. Bioresource Technology, 2013, 145: 25–32
Sun L, Müller B, Westerholm M, Schnürer A. Syntrophic acetate oxidation in industrial CSTR biogas digesters. Journal of Biotechnology, 2014, 171: 39–44
Sahlström L. A review of survival of pathogenic bacteria in organic waste used in biogas plants. Bioresource Technology, 2003, 87(2): 161–166
Chen Y, Fu B, Wang Y, Jiang Q, Liu H. Reactor performance and bacterial pathogen removal in response to sludge retention time in a mesophilic anaerobic digester treating sewage sludge. Bioresource Technology, 2012, 106: 20–26
Acknowledgements
Funding for this study was provided by the Green Farm project supported by a Science Foundation Ireland Investigator Project Award (Ref: 12/IP/1519).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Dennehy, C., Lawlor, P.G., Gardiner, G.E. et al. Process stability and microbial community composition in pig manure and food waste anaerobic co-digesters operated at low HRTs. Front. Environ. Sci. Eng. 11, 4 (2017). https://doi.org/10.1007/s11783-017-0923-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11783-017-0923-9